Abstract
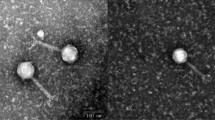
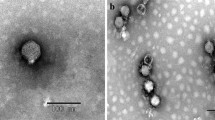
Virulent Cronobacter sakazakii bacteriophage ES2 was isolated from swine fecal samples, and the genome sequence by was determined GS-Flx. Bacteriophage ES2 had a double-stranded DNA genome with a length of 22,162 bp and a G+C content of 50.08%. The morphological characteristics under a transmission electron microscope indicated that bacteriophage ES2 belongs to the family Myoviridae. The structural proteins, including the phage coat protein, were separated by SDS-PAGE and identified by Q-TOF. Bioinformatics analysis of the bacteriophage genome revealed 30 putative open reading frames (ORFs). The predicted protein products of the ORFs were determined and described. To our knowledge, the genome of the newly isolated bacteriophage ES2 was not significantly similar to that of any previously reported bacteriophages of members of the family Enterobacteriaceae.


Similar content being viewed by others
References
Ackermann HW (2003) Bacteriophage observations and evolution. Res Microbiol 154:245–251
Agostoni C, Axelsson I, Goulet O, Koletzko B, Michaelsen KF, Puntis JWL, Rigo J, Shamir R, Szajewski H, Turck D, Vandenplas Y, Weaver LT (2004) Preparation and handling of powdered infant formula: a commentary by the ESPGHAN Committee on nutrition. J Pediatr Gastroenterol Nutr 39:320–322
Biering G, Karlsson S, Clark NC, Jonsdottir KE, Ludvigsson P, Steingrimsson O (1989) Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J Clin Microbiol 27:2054–2056
Borysowski J, Weber-Dabrowska B, Górski A (2006) Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med 231:366–377
Canchaya C, Ventura M, van Sinderen D (2007) Bacteriophage Bioinformatics and Genomics. In: McGrath S, van Sinderen D (eds) Bacteriophage: Genetics and Molecular Biology (Chap. 2). Caister Academic Press, Norfork, UK
Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, Drobniewski F, Lalvani A (2007) A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess 11(3):1–196
Farber JM, Forsythe SJ (2008) Enterobacter sakazakii. ASM Press Inc, Washington
Farmer JJ, Asbury MA, Hickman FW, Brenner DJ (1980) The Enterobacteriaceae Study Group (USA): Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int J Syst Bacteriol 30:569–584
Friedemann M (2007) Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 116:1–10
Greer GG (2005) Bacteriophage control of foodborne bacteria. J Food Prot 68:1102–1111
Hendrix RW (2003) Bacteriophage genomics. Curr Opin Microbiol 6:506–511
Hudson JA, Billington C, Carey-Smith G, Greening G (2005) Bacteriophages as biocontrol agents in food. J Food Prot 68:426–437
Kim KP, Klumpp J, Loessner MJ (2007) Enterobacter sakazakii bacteriophages can prevent bacterial growth in reconstituted infant formula. Int J Food Microbiol 115:195–203
Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, Fronick C, Minx P, Kyung K, Warren W, Fulton R, Feng D, Wollam A, Shah N, Bhonagiri V, Nash WE, Hallsworth-Pepin K, Wilson RK, McClelland M, Forsythe SJ (2010) Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556
Lee SY, Jin HH (2008) Inhibitory activity of natural antimicrobial compounds alone or in combination with nisin against Enterobacter sakazakii. Lett Appl Microbiol 47:315–321
Lee YD, Chang HI, Park JH (2011) Complete genomic sequence of virulent Cronobacter sakazakii phage ESSI-2 isolated from swine feces. Arch Virol 156:721–724
Lu TK, Collins JJ (2007) Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA 104:11197–11202
Madsen SM, Mills D, Djordjevic G, Israelsen H, Klaenhammer TR (2001) Analysis of the genetic switch and replication region of a P335-type bacteriophage with an obligate lytic lifestyle on Lactococcus lactis. Appl Environ Microbiol 67:1128–1139
Manfioletti G, Schneider C (1988) A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res 16:2873–2884
NACMCF (National Advisory Committee on Microbiological Criteria of Foods) (1999) Microbiological evaluations and recommendations on sprouted seeds. Int J Food Microbiol 52:123–153
Nazarowec-White M, Faber JM (1997) Enterobacter sakazakii: a review. Int J Food Microbiol 34:103–113
Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, Jiang W, Chiu W, Hatfull GF, Hendrix RW, King J (2007) Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J Mol Biol 368:966–981
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Sulakvelidze A, Alavidze Z, Morris JG (2001) Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659
Trotter M, McAuliffe O, Callanan M, Edwards R, Fitzgerald GF, Coffey A, Ross RP (2006) Genome analysis of the obligately lytic bacteriophage 4268 of Lactococcus lactis provides insight into its adaptable nature. Gene 366:189–199
Withey S, Cartmell E, Avery LM, Stephenson T (2005) Bacteriophages—potential for application in wastewater treatment processes. Sci Total Environ 339:1–18
Wood WB, Conley MP, Lyle HL, Dickson RC (1978) Attachment of tail fibers in bacteriophage T4 assembly. Purification, properties, and site of action of the accessory protein coded by gene 63. J Biol Chem 253:2437–2445
Acknowledgments
This research was supported financially by Korea Institute of Planning and Evolution for Biotechnology of Food, Agriculture, Forestry and Fisheries funded by Korean Government (108147-02-1-SB010).
Author information
Authors and Affiliations
Corresponding authors
Additional information
J.-H. Park and H.-I. Chang contributed equally to this work.
The complete genome sequence of bacteriophage ES2 was deposited in the GenBank database under Accession Number JF314845.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, YD., Park, JH. & Chang, HI. Genomic sequence analysis of virulent Cronobacter sakazakii bacteriophage ES2. Arch Virol 156, 2105–2108 (2011). https://doi.org/10.1007/s00705-011-1096-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1096-7