Abstract
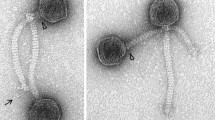
Bacteriophage LF1, a newly isolated temperate phage from a mitomycin-C-induced lysate of wild type Lactobacillus fermentum, was found to contain a double-strand DNA of 42,606 base pairs (bp) with a G+C content of 45%. Bioinformatic analysis of the phage genome revealed 57 putative open reading frames (ORFs). The predicted protein products of ORFs were determined and described. According to morphological analysis by transmission electron microscopy (TEM), LF1 has an isometric head and a non-contractile tail, indicating that it belongs to the family Siphoviridae. The temperate phage LF1 has a good genetic mosaic relationship with ΦPYB5 in the packaging module. To our knowledge, this is first report of genomic sequencing and characterization of temperate phage LF1 from wild-type L. fermentum isolated from Kimchi in Korea.



Similar content being viewed by others
References
Morita H, Yoshkawa H, Sackata R, Nagata Y, Tanaka H (1997) Synthesis of nitric oxide from the two equivalent guanidino nitrogens of l-arginine by Lactobacillus fermentum. J Bacteriol 179(24):7812–7815
Zeng XQ, Pan DD, Zhou PD (2011) Functional characteristics of Lactobacillus fermentum F1. Curr Microbiol 62:27–31
Dickson EM, Riggio MP, Macpherson L (2005) A novel species-specific PCR assay for identifying Lactobacillus fermentum. J Med Microbiol 54:299–303
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Besemer J, Borodovsky M (2005) GeneMark: web software for gene finding in prokaryotes eukaryotes and viruses. Nucl Acids Res 33:W451–W454
Altschul SF, Madden TL, Schaffer AA, Zhang Z, Miller W, Limpman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Darling ACE, Mau B, Blatter FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14(7):1394–1403
Calendar R (2006) The bacteriophages, 2nd edn, Oxford University Press, Oxford; New York Microbiol. Mol. Biol. Rev. 67:238–276
Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H (2003) Prophage genomics. Microbiol Mol Biol Rev 67(2):238–276
Krupovic M, Bamford DH (2008) Holin of bacteriophage lambda: structural insights into a membrane lesion. Mol Microbiol 69(4):781–783
Wood WB, Conley MP, Lyle HL, Dickson RC (1978) Attachment of tail fibers in bacteriophage T4 assembly, purification, properties, and site of action of the accessory protein coded by gene 63. J Biol Chem 253(7):2437–2445
Young R (1992) Bacteriophage lysis: mechanism and regulation. Microbiol Rev 56:430–481
Zhang X, Wang S, Guo T, Kong J (2011) Genome analysis of Lactobacillus fermentum temperate bacteriophage ΦPYB5. Int J Food Microbiol 144:400–405
Zhang X, Kong J, Qu Y (2006) Isolation and characterization of a Lactobacillus fermentum temperate bacteriophage from Chinese yogurt. J Appl Microbiol 101:857–863
Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (2002) Virus taxonomy. Elsevier/Academic Press, London
Juhala RJ, Ford ME, Duda RL, Hatfull AYGF, Hendrix RW (2000) Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27–51
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G (2010) Mechanochemistry of viral DNA packaging motor. J Mol Biol 400(2):186–203
Durmaz E, Miller MJ, Azcarate-peril A, Toon SP, Klaenhammer TR (2008) Genome sequence and characteristics of Lrm1, a prophage from industrial Lactobacillus rhamnosus Straun M1. Appl Environ Microb 74(15):4601–4609
Acknowledgments
This work was supported by grant C00903 from the National Research Foundation of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yoon, B.H., Chang, H.I. Complete genomic sequence of the Lactobacillus temperate phage LF1. Arch Virol 156, 1909–1912 (2011). https://doi.org/10.1007/s00705-011-1082-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1082-0