Abstract
Five Australian potyviruses, passion fruit woodiness virus (PWV), passiflora mosaic virus (PaMV), passiflora virus Y, clitoria chlorosis virus (ClCV) and hardenbergia mosaic virus (HarMV), and two introduced potyviruses, bean common mosaic virus (BCMV) and cowpea aphid-borne mosaic virus (CAbMV), were detected in nine wild or cultivated Passiflora and legume species growing in tropical, subtropical or Mediterranean climatic regions of Western Australia. When ClCV (1), PaMV (1), PaVY (8) and PWV (5) isolates were inoculated to 15 plant species, PWV and two PaVY P. foetida isolates infected P. edulis and P. caerulea readily but legumes only occasionally. Another PaVY P. foetida isolate resembled five PaVY legume isolates in infecting legumes readily but not infecting P. edulis. PaMV resembled PaVY legume isolates in legumes but also infected P. edulis. ClCV did not infect P. edulis or P. caerulea and behaved differently from PaVY legume isolates and PaMV when inoculated to two legume species. When complete coat protein (CP) nucleotide (nt) sequences of 33 new isolates were compared with 41 others, PWV (8), HarMV (4), PaMV (1) and ClCV (1) were within a large group of Australian isolates, while PaVY (14), CAbMV (1) and BCMV (3) isolates were in three other groups. Variation among PWV and PaVY isolates was sufficient for division into four clades each (I-IV). A variable block of 56 amino acid residues at the N-terminal region of the CPs of PaMV and ClCV distinguished them from PWV. Comparison of PWV, PaMV and ClCV CP sequences showed that nt identities were both above and below the 76-77% potyvirus species threshold level. This research gives insights into invasion of new hosts by potyviruses at the natural vegetation and cultivated area interface, and illustrates the potential of indigenous viruses to emerge to infect introduced plants.




Similar content being viewed by others
References
Adams MJ, Antoniw JF, Fauquet CM (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol 150:459–479
Albrechtson SE (2006) Testing methods for seed-transmitted viruses: principles and protocols. CABI Publishing, Wallingford
Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 19:535–544
Baker CA, Jones L (2007) A new potyvirus found in Passiflora incence in Florida. Plant Dis 91:227
Buchen-Osmond C, Crabtree K, Gibbs A, McLean G (1988) Viruses of plants in Australia. Australian National University Printing Service, Canberra
Chang CA (1992) Characterization and comparison of passionfruit mottle virus, a newly recognised potyvirus, with passionfruit woodiness virus. Phytopathology 82:1358–1363
Chen J, Chen J, Chen J, Adams MJ (2001) Molecular characterisation of an isolate of Dasheen mosaic virus from Zantedeschia aethiopica in China and comparisons in the genus Potyvirus. Arch Virol 146:1821–1829
Cheng Y, Jones RAC (2000) Biological properties of necrotic and non-necrotic strains of bean yellow mosaic virus in cool season grain legumes. Ann Appl Biol 136:215–227
Cooper JI, Jones RAC (2006) Wild plants and viruses: under-investigated ecosystems. Adv Virus Res 67:1–47
Coutts BA, Jones RAC (2005) Incidence and distribution of viruses infecting cucurbit crops in the Northern Territory and Western Australia. Aust J Agric Res 56:847–858
Coutts BA, Kehoe MA, Jones RAC (2011) Minimising losses caused by Zucchini yellow mosaic virus in vegetable cucurbit crops in tropical, sub-tropical and Mediterranean environments through cultural methods and host resistance. Virus Res (in press)
Coutts BA, Kehoe MA, Webster CG, Wylie SJ, Jones RAC (2010) Potyviruses of legume weeds and Passiflora spp. from Western Australia: biological properties and phylogenetic placement of coat protein sequences. Abstracts of 11th plant virus epidemiology symposium, plant viruses: exploring natural and agricultural ecosystems. Cornell University, USA
Coutts BA, Walsh JA, Jones RAC (2007) Evaluation of resistance to Turnip mosaic virus in Australian Brassica napus genotypes. Aust J Agric Res 58:67–74
Fang GW, Allison RF, Zambolim EM, Maxwell DP, Gilbertson RL (1995) The complete nucleotide sequence and genome organization of bean common mosaic virus (NL3 strain). Virus Res 39:13–23
Fargette D, Konate G, Fauquet C, Muller E, Peterschmitt M, Thresh JM (2006) Molecular ecology and emergence of tropical plant viruses. Annu Rev Phytopathol 44:235–260
Fauquet CM, Mayo MA, Maniloff J, Desselburger U, Ball LA (eds) (2005) Virus taxonomy. Eighth report of the international committee for the nomenclature of viruses. Elsevier, London
Galloway R, Kemp E (1984) Late Cainozoic environments in Australia. In: Archer M, Clayton G (eds) Vertebrate zoogeography and evolution in Australasia. Hesperian Press, Carlisle, pp 82–95
Gibbs AJ, Gower JC (1960) The use of a multiple transfer method in plant virus transmission studies and some statistical points arising in the analysis of results. Ann Appl Biol 48:75–83
Gibbs A, Mackenzie A (1997) A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J Virol Methods 63:9–16
Gibbs A, Mackenzie A, Blanchfield AL, Cross P, Wilson CR, Kitajima E, Nightingale M, Clements M (2000) Viruses of orchids in Australia: their identification, biology and control. Aust Orchid Rev 65:10–21
Gibbs AJ, Mackenzie AM, Wei K-G, Gibbs MJ (2008) The potyviruses of Australia. Arch Virol 153:1411–1420
Gibbs AJ, Ohshima K (2010) Potyviruses and the digital revolution. Ann Rev Phytopathol 48:205–223
Gibbs AJ, Ohshima K, Phillips MJ, Gibbs MJ (2008b) The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLos One 3(6):e2523. doi:10.1371/journal.pone.0002532
Gibbs AJ, Trueman JWH, Gibbs MJ (2008) The bean common mosaic virus lineage of potyviruses: where did it arise and when? Arch Virol 153:2177–2187
Hopper SD (1979) Biogeographical aspects of speciation in the southwest Australian flora. Annu Rev Ecol Evol Syst 10:399–422
Hopper SD, Gioia P (2004) The southwest Australian floristic region: evolution and conservation of a global hot spot of biodiversity. Annu Rev Ecol Evol Syst 35:623–650
Iwai H, Terahara R, Yamashita Y, Ueda S, Nakamura M (2006) Complete nucleotide sequence of the genomic RNA of an Amami-O-shima strain of East Asian passiflora potyvirus. Arch Virol 151:1457–1460
Jones RAC (1994) Effects of mulching with cereal straw and row spacing on spread of bean yellow mosaic potyvirus into narrow-leafed lupins (Lupinus angustifolius). Ann Appl Biol 124:45–58
Jones RAC (1996) Virus diseases of Australian pastures. In: Chackraborty S, Leath KT, Skipp RA, Pederson GA, Bray RA, Latch GCM, Nutter FW (eds) Pasture and forage crop pathology. American Society of Agronomy, Madison, pp 303–321
Jones RAC (2005) Patterns of spread of two non-persistently aphid-borne viruses in lupin stands under four different infection scenarios. Ann Appl Biol 146:337–350
Jones RAC (2006) Control of plant virus diseases. Adv Virus Res 67:205–244
Jones RAC (2009) Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res 141:113–130
Jones RAC, Coutts BA, Cheng Y (2003) Yield limiting potential of necrotic and non-necrotic strains of Bean yellow mosaic virus in narrow-leafed lupin (Lupinus angustifolius). Aust J Agric Res 54:849–859
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinforma 9:299–306
Latham LJ, Jones RAC (2001) Incidence of virus infection in experimental plots, commercial crops and seed stocks of cool season crop legumes. Aust J Agric Res 52:397–413
Latham LJ, Jones RAC (2003) Incidence of Celery mosaic virus in celery crops in south-west Australia, and its management using a “celery-free period”. Australas Plant Pathol 32:527–531
Latham LJ, Jones RAC (2004) Carrot virus Y: symptoms, losses, incidence, epidemiology and control. Virus Res 100:89–99
Latham LJ, Smith LJ, Jones RAC (2003) Incidence of three viruses in vegetable brassica plantings and associated wild radish weeds in south-west Australia. Australas Plant Pathol 32:387–391
Lin SS, Hou RF, Yeh SD (2001) Complete genome sequence and genetic organization of a Taiwan isolate of Zucchini yellow mosaic virus. Bot Bull Acad Sin 42:243–250
Lovisolo O, Hull R, Rosler O (2003) Coevolution of viruses with hosts and vectors and possible paleontology. Adv Virus Res 62:325–379
Mackenzie AM, Nolan M, Wei KJ, Clements MA, Gowanlock D, Wallace BJ, Gibbs AJ (1998) Ceratobium mosaic potyvirus: another virus from orchids. Arch Virol 143:903–914
McKirdy SJ, Coutts BA, Jones RAC (1994) Occurrence of bean yellow mosaic virus in subterranean clover pastures and perennial native legumes. Aust J Agric Res 45:183–194
Meiners JP, Gillaspie AG, Lawson RH, Smith FF (1978) Identification and partial characterization of a strain of bean common mosaic virus from Rhynchosia minima. Phytopathology 68:283–287
Paczkowska G, Chapman AR (2000) The Western Australian flora—a descriptive catalogue. Wildflower Society of Western Australia. Department of Conservation and Land Management, and Botanic Gardens and Parks Authority, Perth
Parry JN, Davis RI, Thomas JE (2004) Passiflora virus Y, a novel virus infecting Passiflora spp. in Australia and the Indonesian Province of Papua. Australas Plant Pathol 33:423–427
Pio-Ribeiro G, Pappu SS, Pappu HR, Andrade GP, Reddy DVR (2000) Occurrence of Cowpea aphid-borne mosaic virus in peanut in Brazil. Plant Dis 84:760–766
Regatieri LJ, Gaspar JO, Belintani P, Yuki VA (2008) Identification and characterization of a potyvirus isolated from siratro plants. J Phytopathol 156:214–216
Saqib M, Jones RAC, Cayford B, Jones MGK (2005) First report of Bean common mosaic virus in Western Australia. Plant Pathol 54:563
Saqib M, Nouri S, Cayford B, Jones RAC, Jones MGK (2010) Genome sequences and phylogenetic placement of two isolates of Bean common mosaic virus from Macroptilium atropurpureum in north-west Australia. Australas Plant Pathol 39:184–191
Sokhandan N, Gillings MR, Bowyer JW (1997) Polymerase chain reaction detection and assessment of genetic variation in New South Wales isolates of passionfruit woodiness potyvirus. Australas Plant Pathol 26:155–164
Spetz C, Taboada AM, Darwich S, Ramsell J, Salazar LF, Valkonen JPT (2003) Molecular resolution of a complex of potyviruses infecting solanaceous crops at the centre of origin in Peru. J Gen Virol 84:2565–2578
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tairo F, Jones RAC, Valkonen JPT (2006) Potyvirus complexes in sweetpotato: occurrence in Australia, serological and molecular resolution, and analysis of the sweet potato virus 2 (SPV2) component. Plant Dis 90:1120–1128
Taylor RH, Greber RS (1973) Passionfruit woodiness virus. CMI/AAB Descriptions of plant viruses, no. 122
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Thresh JM (1980) The origins and epidemiology of some important plant virus diseases. Appl Biol 5:1–65
Torrance L, Pead MT (1986) The application of monoclonal antibodies to routine tests for two plant viruses. In: Jones RAC, Torrance L (eds) Development in applied biology. 1. Development and applications on virus testing. Association of Applied Biologists, Wellesbourne, pp 103–118
van Regenmortel MHV (2000) Introduction to the species concept in virus taxonomy. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM et al (eds) Seventh report of the international committee for taxonomy of viruses. Academic Press, San Diego, pp 3–16
Webster CG, Coutts BA, Jones RAC, Jones MGK, Wylie SJ (2007) Virus impact at the interface of an ancient ecosystem and a recent agroecosystem: studies on three legume-infecting potyviruses in the southwest Australian floristic region. Plant Pathol 56:729–742
Wylie SJ, Coutts BA, Jones MGK, Jones RAC (2008) Phylogenetic analysis of Bean yellow mosaic virus isolates from four continents: relationship between the seven groups found and their hosts and origins. Plant Dis 92:1596–1603
Wylie SJ, Jones MGK (2011) The complete genome sequence of a Passion fruit woodiness virus isolate from Australia determined using deep sequencing and its relationship to other potyviruses. Arch Virol 156:479–482
Wylie SJ, Jones MGK (2011) Characterisation and quantitation of mutant and wild-type genomes of Hardenbergia mosaic virus isolates co-infecting a wild plant of Hardenbergia comptoniana. Arch Virol (in press)
Wylie SJ, Jones RAC (2009) Role of recombination in the evolution of host specialization within Bean yellow mosaic virus. Phytopathology 99:512–518
Wylie SJ, Nouri S, Coutts BA, Jones MGK (2010) Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch Virol 155:1171–1174
Zheng H, Chen J, Chen J, Adams MJ, Hou M (2002) Bean common mosaic virus isolates causing different symptoms in asparagus bean in China differ greatly in the 5′-parts of their genomes. Arch Virol 147:1257–1262
Acknowledgments
We thank all those mentioned in the text for supplying virus isolates, Adrian Gibbs for helpful discussions, and Stuart Vincent and Eva Gajda for help with plant maintenance and ELISA. The Department of Agriculture and Food Western Australia, and Horticulture Australia Ltd provided financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2011_1046_MOESM1_ESM.ppt
Supplementary Fig. 1. Locations where legume and Passiflora species were sampled in Western Australia: map of the Australian continent showing where samples were taken in three different climatic regions: tropical (Broome, Kununurra, Wyndham), sub-tropical (Carnarvon) and Mediterranean (Geraldton, Pemberton, Perth). (Original sites for each isolate in Table 1). (PPT 148 kb)
705_2011_1046_MOESM2_ESM.ppt
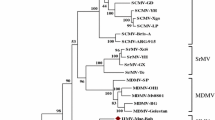
Supplementary Fig. 2. Neighbour-joining relationship phylogram obtained from alignment of 33 complete and seven partial coat protein nucleotide sequences of new potyvirus isolates from wild or cultivated legume and Passiflora spp. and 41 potyvirus sequences from GenBank. The tree was generated using the ClustalW and MEGA 4.1 programmes set to default parameters. Tree branches were bootstrapped with 1,000 replications. Numbers at nodes indicate bootstrap scores of >50%. The scale bar represents a genetic distance of 0.1 for horizontal branch lengths. New sequences are shown in bold without GenBank codes, but earlier sequences are shown with GenBank codes. New partial sequences are shown in bold with grey backgrounds. For isolate designations, see Tables 1 and 2. (PPT 107 kb)
Rights and permissions
About this article
Cite this article
Coutts, B.A., Kehoe, M.A., Webster, C.G. et al. Indigenous and introduced potyviruses of legumes and Passiflora spp. from Australia: biological properties and comparison of coat protein nucleotide sequences. Arch Virol 156, 1757–1774 (2011). https://doi.org/10.1007/s00705-011-1046-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1046-4