Abstract
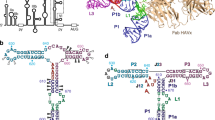
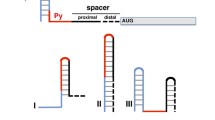
Foot-and-mouth disease virus (FMDV) uses an internal ribosome entry site (IRES), a highly structured segment of its genomic RNA, to hijack the translational apparatus of an infected host. Computational analysis of 162 type II picornavirus IRES RNA sequences yielded secondary structures that included only base pairs supported by comparative or experimental evidence. The deduced helical sections provided the foundation for a hypothetical three-dimensional model of FMDV IRES RNA. The model was further constrained by incorporation of data derived from chemical modification and enzymatic probing of IRES RNAs as well as high-resolution information about IRES RNA-bound proteins.




Similar content being viewed by others
References
Grubman MJ, Baxt B (2004) Foot-and-mouth disease. Clin Microbiol Rev 17:465–493
Mahy BWJ (2005) Introduction and history of foot-and-mouth-disease virus. Curr Top Microbiol Immunol 288:1–8
Clarke BE, Sangar DV, Burroughs JN, Newton SE, Carroll AR, Rowlands DJ (1985) Two initiation sites for foot-and-mouth disease virus polyprotein in vivo. J Gen Virol 66:2615–2626
Belsham GJ (1992) Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J 11:1105–1110
Belsham GJ, Brangwyn JK (1990) A region of the 5’ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol 64:5389–5395
Grubman MJ, Robertson BH, Morgan DO, Moore DM, Dowbenko D (1984) Biochemical map of polypeptides specified by foot-and-mouth disease virus. J Virol 50:579–586
Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62:2636–2643
Kuhn R, Luz N, Beck E (1990) Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J Virol 64:4625–4631
Pelletier J, Sonenberg N (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325
Pilipenko E, Blinov V, Chernov B, Dmitrieva T, Agol V (1989) Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucl Acids Res 17:5701–5711
Robertson BH, Grubman MJ, Weddell GN, Moore DM, Welsh JD, Fischer T, Dowbenko DJ, Yansura DG, Small B, Kleid DG (1985) Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J Virol 54:651–660
Sasaki J, Nakashima N (1999) Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol 73:1219–1226
Deniz N, Lenarcic EM, Landry DM, Thompson SR (2009) Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA 15:932–946
Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T (2007) Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem 282:7770–7776
Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16:2906–2922
Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU (2000) A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev 14:2028–2045
Pacheco A, Martinez-Salas E (2010) Insights into the biology of IRES elements through riboproteomic approaches. J Biomed Biotechnol 2010:458927
Fitzgerald KD, Semler BL (2009) Bridging IRES elements in mRNA to the eukaryotic translational apparatus. Biochim Biophys Acta 1789:518–528
Fernandez-Miragall O, Martinez-Salas E (2003) Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9:1333–1344
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res 31:3406–3415
Boehringer D, Thermann R, Ostareck-Lederer A, Lewis J, Stark H (2005) Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13:1695–1706
Collier A, Gallego J, Klinck R, Cole P, Harris S, Harrison G, Aboul-Ela F, Varani G, Walker S (2002) A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol 9:375–380
Kieft J, Zhou K, Grech A, Jubin R, Doudna J (2002) Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol 9:370–374
Locker N, Easton L, Lukavsky P (2007) HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26:795–805
Lukavsky P, Otto G, Lancaster A, Sarnow P, Puglisi J (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7:1105–1110
Lukavsky P, Kim I, Otto G, Puglisi J (2003) Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10:1033–1038
Pfingsten J, Costantino D, Kieft J (2006) Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314:1450–1454
Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon S, Gorenstein D (2004) Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J Mol Biol 343:805–817
Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM (2006) Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13:1092–1096
Spahn C, Kieft J, Grassucci R, Penczek P, Zhou K, Doudna J, Frank J (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962
Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J (2004) Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118:465–475
Zhao Q, Han Q, Kissinger C, Hermann T, Thompson P (2008) Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr D Biol Crystallogr 64:436–443
Larsen N, Zwieb C (1991) SRP-RNA sequence alignment and secondary structure. Nucl Acids Res 19:209–221
Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF (1980) Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucl Acids Res 8:2275–2293
Zwieb C, Wower I, Wower J (1999) Comparative sequence analysis of tmRNA. Nucl Acids Res 27:2063–2071
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2009) GenBank. Nucl Acids Res 37:D26–D31
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR (2003) Rfam: an RNA family database. Nucl Acids Res 31:439–441
Burks J, Zwieb C, Müller F, Wower I, Wower J (2005) Comparative 3-D modeling of tmRNA. BMC Mol Biol 6:14
Mueller F, Doring T, Erdemir T, Greuer B, Junke N, Osswald M, Rinke-Appel J, Stade K, Thamm S, Brimacombe R (1995) Getting closer to an understanding of the three-dimensional structure of ribosomal RNA. Biochem Cell Biol 73:767–773
Klosterman P, Tamura M, Holbrook S, Brenner S (2002) SCOR: a structural classification of RNA database. Nucl Acids Res 30:392–394
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucl Acids Res 28:235–242
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Gorodkin J, Zwieb C, Knudsen B (2001) Semi-automated update and cleanup of structural RNA alignment databases. Bioinformatics 17:642–645
Andersen E, Lind-Thomsen A, Knudsen B, Kristensen S, Havgaard J, Torarinsson E, Larsen N, Zwieb C, Sestoft P, Kjems J, Gorodkin J (2007) Semiautomated improvement of RNA alignments. RNA 13:1850–1859
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Jang S, Wimmer E (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev 4:1560–1572
Mason PW, Bezborodova SV, Henry TM (2002) Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J Virol 76:9686–9694
Nayak A, Goodfellow IG, Woolaway KE, Birtley J, Curry S, Belsham GJ (2006) Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J Virol 80:9865–9875
Tiley L, King AM, Belsham GJ (2003) The foot-and-mouth disease virus cis-acting replication element (cre) can be complemented in trans within infected cells. J Virol 77:2243–2246
Thiviyanathan V, Yang Y, Kaluarachchi K, Rijnbrand R, Gorenstein DG, Lemon SM (2004) High-resolution structure of a picornaviral internal cis-acting RNA replication element (cre). Proc Natl Acad Sci USA 101:12688–12693
Fernández-Miragall O, López de Quinto S, Martínez-Salas E (2009) Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res 139(2):172–182
Luz N, Beck E (1990) A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett 269:311–314
Luz N, Beck E (1991) Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol 65:6486–6494
Witherell GW, Gil A, Wimmer E (1993) Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry 32:8268–8275
Fernandez-Miragall O, Martinez-Salas E (2007) In vivo footprint of a picornavirus internal ribosome entry site reveals differences in accessibility to specific RNA structural elements. J Gen Virol 88:3053–3062
Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E (2006) Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 12:223–234
Fernández N, García-Sacristán A, Ramajo J, Briones C, Martínez-Salas E (2011) Structural analysis provides insights into the modular organization of picornavirus IRES. Virology 409:251–261
Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM (2005) RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127:4223–4231
Lopez de Quinto S, Martinez-Salas E (1997) Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol 71:4171–4175
Robertson M, Seamons R, Belsham G (1999) A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5:1167–1179
Lopez de Quinto S, Martinez-Salas E (2000) Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380–1392
Lopez de Quinto S, Lafuente E, Martinez-Salas E (2001) IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7:1213–1226
Bassili G, Tzima E, Song Y, Saleh L, Ochs K, Niepmann M (2004) Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J Gen Virol 85:2555–2565
Clark AT, Robertson ME, Conn GL, Belsham GJ (2003) Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol 77(23):12441–12449
Martinez-Salas E (2008) The impact of RNA structure on picornavirus activity. Trends Microbiol 15:230–237
Jucker FM, Pardi A (1995) Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry 34:14416–14427
Hoffman MA, Palmenberg AC (1995) Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol 69:4399–4406
Johansson S, Niklasson B, Maizel J, Gorbalenya AE, Lindberg AM (2002) Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J Virol 76:8920–8930
Kolupaeva V, Pestova T, Hellen C, Shatsky I (1998) Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 273:18599–18604
LaRue R, Myers S, Brewer L, Shaw DP, Brown C, Seal BS, Njenga MK (2003) A wild-type porcine encephalomyocarditis virus containing a short poly(C) tract is pathogenic to mice, pigs, and cynomolgus macaques. J Virol 77:9136–9146
Pevear DC, Luo M, Lipton HL (1988) Three-dimensional model of the capsid proteins of two biologically different Theiler virus strains: clustering of amino acid difference identifies possible locations of immunogenic sites on the virion. Proc Natl Acad Sci USA 85:4496–4500
Kaminski A, Howell MT, Jackson RJ (1990) Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J 9:3753–3759
Sangar DV, Newton SE, Rowlands DJ, Clarke BE (1987) All foot and mouth disease virus serotypes initiate protein synthesis at two separate AUGs. Nucl Acids Res 15:3305–3315
Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN (2007) Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA 13:1366–1374
Niepmann M (1996) Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett 388:39–42
Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM (2000) Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol 20:1583–1595
Kaminski A, Jackson R (1998) The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626–638
Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ (2009) Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell 34:556–568
Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S (2005) The polypyrimidine tract binding protein is a monomer. RNA 11:1803–1808
Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M (2005) Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11:1809–1824
Niepmann M, Petersen A, Meyer K, Beck E (1997) Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol 71:8330–8339
Oh YL, Hahm B, Kim YK, Lee HK, Lee JW, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang SK (1998) Determination of functional domains in polypyrimidine-tract-binding protein. Biochem J 331(Pt 1):169–175
Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S (2004) Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure 12:1631–1643
Kaminski A, Hunt SL, Patton JG, Jackson RJ (1995) Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924–938
Kolupaeva V, Hellen C, Shatsky I (1996) Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199–1212
Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH (2005) Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309:2054–2057
Beales L, Holzenburg A, Rowlands D (2003) Viral internal ribosome entry site structures segregate into two distinct morphologies. J Virol 77:6574–6579
Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S (2007) Structural insights into the transcriptional and translational roles of Ebp1. EMBO J 26:3936–3944
Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE (2000) Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 275:41369–41376
Saleh L, Rust RC, Fullkrug R, Beck E, Bassili G, Ochs K, Niepmann M (2001) Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J Gen Virol 82:757–763
Pestova T, Shatsky I, Hellen C (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 16:6870–6878
LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE (2006) Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281:22917–22932
Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JWB, Doudna JA, Robinson CV, Leary JA (2007) Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics 6:1135–1146
Benne R, Hershey JW (1976) Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc Natl Acad Sci USA 73:3005–3009
Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW (2004) The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem 279:8946–8956
Siridechadilok B, Fraser C, Hall R, Doudna J, Nogales E (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310:1513–1515
Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N (2007) Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucl Acids Res 35:1514–1521
Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K (2001) Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem 276:20824–20826
Tolan DR, Traut RR (1981) Protein topography of the 40 S ribosomal subunit from rabbit reticulocytes shown by cross-linking with 2-iminothiolane. J Biol Chem 256:10129–10136
Ben-Shem A, Jenner L, Yusupova G, Yusupov M (2010) Crystal structure of the eukaryotic ribosome. Science 330:1203–1209
Pfingsten JS, Kieft JS (2008) RNA structure-based ribosome recruitment: lessons from the Dicistroviridae intergenic region IRESes. RNA 14:1255–1263
Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26:41–50
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Acknowledgments
This research was supported by grants from the Alabama Agricultural Experiment Station Foundation to I.K.W. and J.W. and an Auburn University Biogrant to J.W. Publication costs were supported in part by the Upchurch Fund for Excellence. J.M.B. was supported by the National Science Foundation under Grant No. 0091853 and NSF-EPS 0447675. The molecular graphics image in Fig. 4 was produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
FMDVIRES-alignment.fas. Picornavirus type II IRES RNA alignment
162 Sequences from aphthoviruses, cardioviruses and parechoviruses were aligned in fasta format as described in the text (PDB 2340 kb)
FMDV-PTBd34-model.pdb. Model of FMDV IRES RNA complexed with PTB domains III and IV
Coordinates in PDB format of tertiary structure model of FMDV IRES RNA complexed with structure of PTB domains III and IV as described in text (FAS 109 kb)
Rights and permissions
About this article
Cite this article
Burks, J.M., Zwieb, C., Müller, F. et al. In silico analysis of IRES RNAs of foot-and-mouth disease virus and related picornaviruses. Arch Virol 156, 1737–1747 (2011). https://doi.org/10.1007/s00705-011-1043-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1043-7