Abstract
Sequence analysis of the nucleoprotein (NP) of swine-origin influenza virus H1N1 (S-OIV) reveals a number of atypical characteristics including an early start codon and a highly conserved, non-aromatic residue at position 313. Using an in vitro viral polymerase reconstitution assay, we found that the polymerase complex containing the NP of S-OIV (NPS-OIV) yielded substantially lower activity than those assayed with NP derived from other influenza virus strains. Moreover, alteration of the early start codon or introduction of an aromatic residue at position 313 (V313Y) did not increase but instead exacerbated the poor polymerase activity. Interestingly, when NPS-OIV was allowed to compete with that of a mouse-adapted influenza virus (A/PR/8/34) to form progeny virions, only progeny bearing NPS-OIV were produced, despite the low polymerase activity associated with NPS-OIV. Our results indicated that NPS-OIV requires both the early start codon and the V313 residue for its optimal function. These characteristics are required for a strong compatibility between the S-OIV polymerase subunits and its indigenous NP over that of other strains, which might explain why productive reassortment between S-OIV and seasonal influenza viruses has yet to occur in nature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although considered relatively mild, the pandemic swine-origin influenza virus H1N1 (S-OIV) has been known to cause severe disease in certain groups of individuals, such as young children, pregnant women and those with an underlying lung condition [1–3]. Like other seasonal influenza viruses, S-OIV has been reported to transmit rapidly among humans. Nevertheless, the molecular mechanism underlying this phenomenon has not been clearly understood. Characteristics known to be associated with human adaptation, such as E627K and D701N mutation of the PB2 polymerase subunit, are not found in S-OIV [4–6]. Recently, the presence of a basic residue at position 591 (R591 or K591) of the PB2 protein of S-OIV has been proposed to be associated with its high transmissibility in mammals [7, 8], suggesting that the polymerase complex of S-OIV might be responsible for the high transmissibility of the virus.
Given the fact that S-OIV has been co-circulating worldwide with seasonal human influenza viruses H1N1 and H3N2 or highly pathogenic avian influenza virus H5N1, it is feared that reassortment between S-OIV and other unrelated viral strains might take place, giving rise to a new reassortant with a pandemic potential. Surprisingly, despite more than a year of circulation in humans, there has been no report of reassortment between S-OIV and other influenza viruses in humans. The recent study of Vijaykrishna et al. [9] has reported reassortment between S-OIV and another swine influenza virus in pigs. However, only one variant was identified; its genetic constellation was found to harbor only the NA segment of S-OIV, while other segments, especially those of the polymerase complex, still belong to the triple-reassortant virus.
The influenza virus polymerase complex, comprising PB2, PB1 and PA subunits, plays critical roles in viral replication, especially in steps involving RNA replication and transcription. The subunits form heterotrimeric complexes within virus particles or in the nuclei of infected cells. The function of the heterotrimeric complex depends largely on its assembly into a ribonucleoprotein particle (RNP), in which the vRNA segment is associated with the nucleoprotein (NP) and the three polymerase subunits. Originally derived from a classical swine influenza virus, the NP of S-OIV (NPS-OIV) still possesses some characteristics that are not commonly found in viruses of human or avian origin. The NP segment of the majority of S-OIV isolates (>99%) harbors adenosine instead of cytosine at position 28 in the 3’ untranslated region (UTR), resulting in an early start codon and consequently six extra amino acids at the N terminus. Moreover, a highly conserved residue at position 313, which typically bears an aromatic side chain – tyrosine (Y313) for human and phenylalanine (F313) for avian and swine influenza viruses – has been substituted by valine in virtually all S-OIV isolates [10, 11]. Whether these uncommon characteristics of NPS-OIV have any functional implications has never been explored.
In this study, we demonstrated that the early start codon and the presence of a non-aromatic residue are crucial for the compatibility of NPS-OIV with the polymerase complex of S-OIV and provided evidence indicating that selective compatibility between the NP protein and the polymerase complex of S-OIV might contribute, at least in part, to the restriction of reassortment with those of other influenza viruses.
Materials and methods
Cells and viruses
Madin-Darby canine kidney (MDCK) and human embryonated kidney (HEK293T) cells were cultured in Opti-MEM medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics. Cell cultures were maintained at 37°C in a humidified 5% CO2 incubator. Unless otherwise indicated, influenza viruses were grown in MDCK cells or 10-day-old embryonated chicken eggs for 48 h at 37°C. Culture supernatants or allantoic fluids were collected and cleared by centrifugation at 1,000 x g for 10 min before determining the presence of virus by the hemagglutination assay using type-O human red blood cells.
Plasmid construction
The construction of pHW2000 plasmids expressing all eight genes of S-OIV were carried out as described previously [12]. Briefly, full-length cDNA of each gene was generated by RT-PCR (TAKARA) from viral RNA extracted from egg-derived A/Nonthaburi/102/09 (H1N1) using universal primer pairs as described [13]. The PCR products were digested with BsmBI or BsaI before cloning into BsmBI-digested pHW2000 plasmid. The plasmids that were obtained were subjected to nucleotide sequencing to ensure that no unwanted mutations were present. Site-specific NP mutations were performed by PCR of pHW2000 expressing NP of PR8 or S-OIV with mutagenic primers designed to include the desired mutations. After PCR amplification by Hi-fidelity DNA polymerase (KAPA Biosystems), the reaction mixture was digested with DpnI and subsequently used to transform competent cells.
The plasmid expressing EGFP-NP fusion protein (pEGFP-NP) was constructed by first introducing an MluI restriction site into the 3’-UTR region of plasmids expressing each variant of the NP gene by PCR-based site directed mutagenesis. The full-length EGFP was then amplified from the pCruz-EGFP plasmid (Santacruz Biotechnology) using the primers 5’- TATAACGCGTACCATGGGATCCGTGAGCAA-3’ and 5’- TATAACGCGTGAATTCCTTGTACAGCTCGTC-3’ (MluI restriction site underlined). The modified plasmid and PCR product were digested with MluI and ligated together, giving rise to a plasmid expressing EGFP fused in-frame with the NP protein. All of the plasmids expressing fusion proteins were verified by sequencing before they were used in subsequent experiments.
The construction of the reporter plasmid, designated PolI-sNA-GFP, was carried out by replacing the ORF of segment 7 of A/PR/8/34 expressed in the backbone of a bidirectional plasmid, pHW2000, with a combination of a globular head region of NA of A/Vietnam/DT/036 (H5N1) engineered to contain the murine Ig κ chain signal peptide sequence (METDTLLLWVLLLWVPGSTGD) followed by IRES of EMCV and GFP, resulting in the plasmid pHW-sNA-GFP. Consequently, the sNA-IRES-GFP transcript was flanked by the 3’UTR and 5’UTR of segment 7 of influenza A virus. The CMV promoter of plasmid pHW-sNA-GFP was then removed to allow only expression of negative-sense RNA from a human RNA polymerase I promoter, giving rise to the final construct, PolI-sNA-GFP. The PolI-driven Ds-Red plasmid was constructed using the same strategy.
Reverse genetics of influenza virus
Construction of recombinant influenza viruses was carried out by 8-plasmid transfection following previously described protocols [12, 14]. Briefly, co-cultures of HEK293T-MDCK cells were transfected with plasmids expressing each gene of influenza virus using TransIT-LT1 (Mirus Corporation). Cells were incubated with transfection mixture for 8 h before being washed and replaced with fresh Opti-MEM medium. At 24 h post-transfection, 2.0 μg/ml of TPCK-treated trypsin (Sigma-Aldrich) was added into the co-culture. Once a cytopathic effect was observed, the supernatant was harvested, inoculated into 10-day-old embryonated chicken eggs, and incubated at 37°C for 48 h. Allantoic fluids were harvested and subjected to hemagglutination assay to determine the presence of the virus. Total RNA of progeny viruses generated from plasmids bearing specific mutations was extracted and subjected to RT-PCR followed by nucleotide sequencing to confirm the presence of mutations.
Influenza virus polymerase reconstitution assay
The experiments were carried out according to Wanitchang et al. [15] with slight modifications. In brief, HEK293T cells (1 × 106 cells) were transfected with a plasmid mixture containing either PR8- or S-OIV-derived PB2-,PB1-, PA- and NP-expression plasmids as well as the PolI-sNA-GFP as a reporter plasmid. The transfection mixture also contained a plasmid expressing Ds-Red to normalize for variation in transfection efficiency. The polymerase activity was assessed 48 h post-transfection by visually monitoring the expression of GFP using fluorescent microscopy or by quantifying the NA activity in the supernatant using 2’-(-4methylumbelliferyl)-α-D-N-acetylneuraminic acid (MU-NANA) as described previously [16]. For assays using EGFP-NP fusion protein, a PolI-driven plasmid transcribing DsRed (PolI-DsRed) was used as a reporter plasmid, and analysis was performed by fluorescent microscopy.
NP competition assay
HEK293T cells (5 × 106 cells) were trasfected with various amounts of pHW2000 plasmid expressing the NP of PR8 using Fugene HD transfection reagent (Roche). At 12 h post-transfection, S-OIV was allowed to adsorb to transfected cells (MOI = 0.01) for 1 h at 37°C before the culture was washed and the medium replaced with OptiMEM medium containing 2 μg TPCK-trypsin. At 24 h postinfection, cell supernatants were then harvested and used to infect the MDCK monalayer until CPE was observed. Culture supernatants were subjected to viral RNA extraction. An equal amount of total RNA (100 ng) was treated with DNase-I and used for amplification of the NP fragment by one-step RT-PCR (TAKARA) using a primer pair specific for the NP of S-OIV or PR8 that was designed to distinguish each RNA transcript. The sequences of primers are available upon request. As an internal control, primers specific for the HA of S-OIV were used to normalize for the amount of RNA template.
Western blotting
HEK293T cells, transfected as described for the polymerase reconstitution assay, were lysed in mammalian cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40). The extracts were separated by SDS-PAGE and blotted onto the nitrocellulose membranes (Amersham). The membranes were blocked for 1 h in TBS containing 5% milk, followed by incubation with rabbit anti-NP (eEnzyme) or rabbit anti-GFP (Santacruz Biotechnology) for 1 h. After several washes, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibodies (Santacruz Biotechnology) for 1 h and developed using a SuperSignal West Pico kit (Thermo Scientific) according to the manufacturer’s instructions.
Results
Unique characteristics of NPS-OIV are required for its optimized function in polymerase assays with the polymerase complex of S-OIV
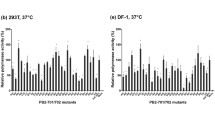
We first assessed the activity of the polymerase complex in which NP derived from various strains of influenza viruses, i.e., A/PR/8/34 (PR8; Genbank EF190983.1), A/Brisbane/59/2007 (Seasonal H1N1; Genbank CY064975.1), A/Nonthaburi/102/09 (S-OIV; Genbank GQ166232.1), and A/open-billed stork/Bangkok/LBD0511F/04 (H5N1; Genbank EF112294.1), were complexed with three polymerase subunits derived from PR8 or S-OIV. As shown in Fig. 1, the level of polymerase activity of the RNP bearing NPS-OIV was substantially lower than that of RNPs complexed with NPs of other strains. Surprisingly, even though the polymerase subunits of S-OIV were assayed with NPS-OIV, the resulting polymerase activity was still lower than that observed with the other NPs tested in this study.
Polymerase activity of RNP of PR8 and S-OIV assayed with different NP proteins. HEK293T cells were transfected with plasmid pHW2000 expressing NP of S-OIV, PR8, seasonal influenza virus H1N1 (sH1N1) or avian influenza H5N1 together with those expressing PB2, PB1 and PA of S-OIV or PR8, using PolI-sNA-GFP as a reporter plasmid. Transfected cells were cultured at 37°C for 48 h. The supernatants were harvested, and the NA activity was determined. Polymerase activity was calculated relative to that obtained from assays using NP derived from the same source of the three polymerase subunits. The results shown are the average of at least three independent experiments. Error bars represent standard deviation
To determine whether the extra amino acids of NPS-OIV resulting from an early start codon play any role in the polymerase activity, we performed site-directed mutagenesis to change the first ATG to CTG and subsequently assessed polymerase activity when this NP was complexed with the polymerase subunits of S-OIV or PR8. Moreover, we altered the second ATG to CTG to determine if the first start codon was required for optimal function of NPS-OIV (Fig. 2A). As a control, we also constructed an S-OIV-like version of NPPR8 by introducing an early start codon in the 3’-UTR of NPPR8 (Fig. 2A). Interestingly, the absence of the early start codon in the 3’-UTR of NPS-OIV resulted in a decrease in polymerase activity relative to wild-type NPS-OIV (Fig. 2B). However, the polymerase activity of NPS-OIV with the early start codon alone was notably higher than that of wild-type NPS-OIV (Fig. 2B). It is also important to note that the absence of the early start codon showed only a marginal effect on the polymerase activity when assayed with the polymerase subunits of PR8 (Fig. 2B). Of note, the presence of an early start codon in NPPR8 did not appear to affect the overall polymerase activity in the same manner observed with NPS-OIV. In fact, introduction of an early start codon in NPPR8 resulted in a slight decrease in polymerase activity when tested with the polymerase subunits of both PR8 and S-OIV (Fig. 2C). Taken together, these data suggest that the early start codon of NPS-OIV, or the presence of the extra N-terminal amino acids, is required for the optimal function of the protein when specifically assembled with the polymerase subunits of S-OIV.
The early start codon at the 3’UTR of NP S-OIV is required for its optimal function when assayed with the polymerase complex of S-OIV. (A) Schematic representation of the NP segment showing differences in the 3’-UTRs of PR8 and S-OIV. Start codons are underlined. (B, C) pHW2000 expressing NPS-OIV or NPPR8 was modified to harbor only the first or second start codon and subsequently tested in the polymerase reconstitution assay together with the polymerase complex of PR8 or S-OIV. Polymerase activity was determined based on the NA activity in the supernatants of transfected cells and calculated relative to those obtained using wild-type NP. The results shown are the average of at least three independent experiments, and error bars represent standard deviation. (D) pHW2000 expressing NPS-OIV was modified to shift the first start codon to the +1 position (NPS-OIV+1) and subsequently used to measure the polymerase activity with polymerase subunits of PR8 or S-OIV in comparison to wild-type NPS-OIV. Polymerase activity was determined based on the NA activity in the supernatants of transfected cells and calculated relative to that obtained from assays using wild-type NPS-OIV. The results shown are the average of at least three independent experiments. Error bars represent standard deviation
It is notable that the position of the early start codon in the 3’-UTR of NPS-OIV does not meet the criteria of a strong Kozak consensus sequence [17]. As shown in Fig. 2A, the early start codon has T at nucleotide position +4 and A at position -3, which is in contrast with the second start codon, which contains G at both positions +4 and -3. It is thus likely that leaky ribosomal scanning would occur to bypass the early start codon, giving rise to a mixture of wild-type NP and the N-terminally extended species, analogous to the translation of various proteins from PB1 mRNA reported previously [18]. To address this hypothesis, we modified the early start codon by shifting the reading frame to the +1 position to create an altered amino acid sequence for this ORF (NPS-OIV+1) and determined the effect of NP function in comparison with the wild-type NPS-OIV using the polymerase reconstitution assay. As shown in Fig. 2D, when assayed with the polymerase subunits derived from PR8, there was no significant difference in the polymerase activity generated from the two NPS-OIV constructs. However, when NPS-OIV+1 was assayed with the polymerase proteins of S-OIV, the polymerase activity was substantially lower than when using wild-type NPS-OIV (Fig. 2D). These data indicate that translation of NPS-OIV mRNA gives rise to a mixture of long and normal-size proteins, presumably through leaky ribosomal scanning process. Moreover, in line with results presented in Fig. 2B, the presence of NPS-OIV with extra N-terminal amino acids could augment the polymerase activity of the S-OIV polymerase complex.
We next sought to examine the functional implications of the highly conserved residue at position 313 of NPS-OIV. We altered valine at position 313 of NPS-OIV to tyrosine (V313Y) as well as tyrosine at the same position of NPPR8 to valine (Y313V) and subsequently measured the polymerase activity assayed with polymerase proteins of PR8 or S-OIV. As shown in Fig. 3A and B, it is clear that mutation of residue 313 of both NPPR8 and NPS-OIV could substantially affect the polymerase activity of RNP of PR8 and S-OIV. While the Y313V mutation of NPPR8 augmented the polymerase activity of S-OIV RNP more than twofold, this particular variant, when assayed with the polymerase subunits of PR8, resulted in a sharp reduction in polymerase activity (Fig. 3A). Moreover, the V313Y mutation of NPS-OIV was found to substantially impair the function of NPS-OIV when assembled with the polymerase complex of S-OIV. However, this decrease in activity was not observed when it was tested with the polymerase complex of PR8 (Fig. 3B). Of note, we cannot rule out the possibility that the altered NP function observed in the polymerase reconstitution assay of each NP variant was due to the compromised stability of the mutant protein. To investigate this issue, we performed polymerase reconstitution assays using PB2, PB1 and PA of PR8 and all variants of NP proteins including NPPR8, NPPR8-Y313V, NPS-OIV, NPS-OIV-V313Y, NPS-OIV-ΔATG1 and NPS-OIV-ΔATG2 using PolI-sNA-GFP plasmid as a reporter and assessed the expression of the NP protein as well as GFP by western blot analysis. As depicted in Fig. 4A, while commercially available polyclonal antibodies raised against the NP of influenza A virus strongly reacted with NPPR8 and its mutant, they were unable to react with any of the variants of NPS-OIV. Since the bands of GFP could be detected in the blot, we believe that failure to detect the band of NPS-OIV was due to antigen-antibody incompatibility rather than loss of protein stability. To explore this issue further, we constructed a fusion protein in which EGFP was fused in-frame with the N-terminal region of all of the NP variants. When HEK293T cells were transfected with plasmids expressing the EGFP-NP fusion protein, we observed comparable EGFP expression level with all constructs (Fig. 4B). Of note, EGFP was found to be predominantly expressed in the nuclei of transfected cells, which is consistent with the localization of the NP protein reported elsewhere [19, 20]. To our surprise, we found that the EGFP-NP fusion proteins were functionally active in the polymerase reconstitution assay. Using the PolI-driven DsRed plasmid as the reporter, we observed comparable expression of EGFP but substantially different DsRed expression when NPS-OIV and NPS-OIV-V313Y were assayed with the polymerase proteins of S-OIV (Fig. 4C), indicating that the alteration of amino acid position 313 and the presence of extra amino acids at the N terminus have no effect on the stability of the NP protein, be it of PR8 or S-OIV origin. Collectively, these results demonstrate the highly specific and strong compatibility of polymerase proteins with NPS-OIV. Furthermore, the presence of a non-aromatic amino acid, most likely valine, at position 313 of NPS-OIV is necessary for the RNP of S-OIV to function optimally.
The valine residue at position 313 of NP S-OIV is critical for optimal function when assayed with the polymerase complex of S-OIV. (A) pHW2000 expressing NPPR8 and (B) pHW2000 expressing NPS-OIV were subjected to site-directed mutagenesis to modify the conserved amino acid residue at position 313, Y313V for NPPR8 and V313Y for NPS-OIV. Subsequently, the modified plasmid was tested in the polymerase reconstitution assay in combination with PB2, PB1 and PA subunits of PR8 or S-OIV. Polymerase activity was determined based on the NA activity in the supernatants of transfected cells and calculated relative to those obtained from assays using wild-type NPPR8 or NPS-OIV. The results shown are the average of at least three independent experiments. Error bars represent standard deviation
Changes in the function of mutant NP is not due to loss of protein stability. (A) pHW2000 plasmids expressing wild-type NPPR8 and NPS-OIV and other variants including NPPR8, NPPR8-Y313V, NPS-OIV, NPS-OIV-V313Y, NPS-OIV-ΔATG1 and NPS-OIV-ΔATG2 were assayed with the polymerase subunit of PR8 in the polymerase reconstitution assay using PolI-sNA-GFP as reporter. At 48 h post-transfection, cells were lysed and subjected to western blot analysis using rabbit polyclonal antibodies against influenza A NP and GFP proteins for detecting. (B) pHW2000 plasmids expressing EGFP-NP fusion proteins of wild-type and mutant NPPR8 and NPS-OIV were constructed as described in Materials and methods and used to transfect HEK293T cells. At 24 h post-transfection, transfected cells were examined by fluorescent microscopy. Images were taken at 40X magnification. (C) pHW2000 plasmids expressing EGFP-NP fusion proteins of wild-type NPS-OIV and NPS-OIV-V313Y were assayed with polymerase proteins of S-OIV using PolI-driven DsRed as reporter plasmid. At 24 h after transfection, transfected cells were examined by fluorescent microscopy. Images were taken at 40X magnification
NPS-OIV limits reassortment between S-OIV and another influenza virus
Given that NPS-OIV has unique characteristics that were never found in other NPs of human influenza viruses and that reassortment between S-OIV and other human influenza viruses has never been reported, it is possible that these characteristics might serve as a restriction factor limiting the chance of S-OIV reassorting with other influenza virus strains. To address this hypothesis, we first attempted to use reverse genetics to construct a recombinant influenza virus, designated S-OIVNP-PR8, in which seven genes were derived from S-OIV and the NP was from PR8. Despite a successful rescue of wild-type S-OIV, no productively growing S-OIVNP-PR8 could be rescued either from cells or embryonated eggs in at least three independent experiments (data not shown). These results suggest that the NPPR8 might impair the growth of S-OIV. To further analyze whether the compatibility of NP and the polymerase complex of S-OIV was responsible for growth restriction of the reassortant virus, we first transfected HEK293T cells with an excess amount of plasmid expressing NPPR8 followed by infection of transfected cells by wild-type S-OIV. Progeny virions were subsequently propagated in MDCK cells for at least three passages to ensure that the resulting progeny represented those with the fittest phenotype. As expected, most, if not all, progeny virions contained NPS-OIV (Fig. 5A). Notably, reverse-genetics-derived PR8 virus bearing NPS-OIV could be rescued, but compared with the wild-type PR8, it showed retarded growth characteristics both in MDCK cells and embryonated chicken eggs (Fig. 5B). Collectively, these data indicate that a strong and highly specific compatibility between NP and the polymerase complex of S-OIV plays a crucial role in restricting viral reassortment in infected cells.
NP S-OIV is required for S-OIV growth. (A) HEK293T cells were transfected with various concentrations of pHW2000 expressing NPPR8 (1, 1.5 and 2 μg). After transfection, S-OIV was allowed to infect transfected cells (MOI = 0.01), and progeny virions were further propagated in MDCK cells for three passages. Cell supernatants were subjected to viral RNA extraction. Equal amounts of DNase-I treated viral RNA was analyzed by RT-PCR analysis using primers specific for NPPR8, NPS-OIV and HAS-OIV. (B) Reverse genetics was carried out to construct a recombinant PR8 bearing NPS-OIV. Growth of recombinant virus (7PR8+NP(S-OIV)) was compared with wild-type PR8 in both embryonated chicken eggs (n=10) and MDCK cells. Viral titers were determined after 48 h incubation in eggs or cells by TCID50 assay in MDCK cells. Error bars represent standard deviation of three independent assays
Discussion
When two or more different influenza viruses infect the same cell, their segmented genomes can undergo reassortment, which, as a result, generates progeny virions bearing different combinations of gene segments. Indeed, the pandemic influenza viruses of 1957, 1968 and 2009 emerged through reassortment of genes derived from different influenza viruses [21, 22]. Interestingly, since its introduction to the human population in March 2009, S-OIV, in spite of its highly efficient human-to-human transmission, has not been reported to reassort with other human influenza viruses. This observation may have been due to unfavorable characteristics of reassortant viruses, that is, their replication efficiency in humans may be somehow restricted by the specific genetic constellation of S-OIV. Accumulating evidence has indicated that compatibility between influenza polymerase complexes plays a pivotal role in influenza virus reassortment [23, 24]. Indeed, a recent study by Octaviani and colleagues demonstrated that compatibility between the polymerase complex of S-OIV and H5N1 virus is critical for generation of reassortant viruses in vitro [25]. Here, we have characterized NPS-OIV, a critical protein of the polymerase complex of S-OIV, and showed that it possesses a number of unique characteristics that may, at least in part, contribute to the limited compatibility between the polymerase complex of S-OIV and those of other strains.
It is interesting to note that results from in vitro polymerase reconstitution assays performed to determine the activity of polymerase subunits of S-OIV with NP derived from various other influenza viruses revealed that the activity of the RNP bearing NPS-OIV was the lowest among all of the ones tested. However, only NPS-OIV can be used to rescue recombinant S-OIV in vitro. Data from our reverse genetics and competition assays between NPPR8 and NPS-OIV also showed that virtually all of the S-OIV progeny contained NPS-OIV. In contrast, while NPS-OIV could be used to generate recombinant virus in the backbone of PR8, the resulting recombinant virus clearly showed retarded growth characteristics both in MDCK cells and embryonated eggs compared to wild-type PR8. Collectively, these data indicate that NPS-OIV is required for optimal growth of S-OIV despite the low level of polymerase activity of the S-OIV polymerase complex.
At least two unique attributes of NPS-OIV might be responsible for the highly selective compatibility with the S-OIV polymerase complex. The majority of NPS-OIV possessed an atypical start codon in the 3’ UTR of segment 5 of the viral genome. As a result, NPS-OIV is expected to possess six additional amino acids at its N terminus. Our results also indicated that the presence of the early start codon is more favorable than that of the variant in which the start codon was changed to the one typically found in most NPs. However, when NPPR8 was modified to harbor the early start codon, the level of polymerase activity was substantially reduced. These data thus strongly indicate that the favorable effect of the early start codon of NP is specific for NP and the polymerase complex of S-OIV. Whether the extra amino acids in the N-terminal region of NPS-OIV have any functional role is not clear. Since the N-terminal region of all NP proteins harbors a strong nuclear localization signal and domains believed to interact with cellular proteins [26], we speculate that additional amino acids in this region might affect these properties of NPS-OIV.
While the majority of NPS-OIV proteins possess valine at position 313 (V313), those of virtually all influenza viruses harbor aromatic residues, either tyrosine or phenylalanine. We demonstrated that the presence of V313 is critical for NPS-OIV to function optimally with the polymerase complex of S-OIV. To our surprise, when tyrosine at position 313 of NPPR8 was substituted by valine (Y313V), the polymerase activity, when assayed with polymerase complex of S-OIV, was markedly elevated compared to wild-type NPPR8. In addition, the alteration of V313 of NPS-OIV to Y313 (V313Y) resulted in a substantial reduction in the activity of the S-OIV polymerase complex. Notably, both the Y313V mutation of NPPR8 and the V313Y mutation of NPS-OIV resulted in a decrease in activity of the PR8 polymerase complex. These results clearly indicate that the presence of V313 in NPS-OIV did not occur by chance but in order to enhance the compatibility between NP and the polymerase complex of S-OIV. Thus far, the functional implications of V313 of S-OIV have not been clearly established. Based on the crystal structure of the NP protein, the side chain of residue 313 is likely to be exposed on the surface of the protein [27]. This opens the possibility that this site may be crucial for the direct contact between NP and other proteins of the polymerase complex. Biochemical evidence from previous studies would predict that either PB2 or PB1 might be a candidate [28–30], but further investigation is necessary to prove this hypothesis. It is also interesting to note that several amino acids in nearby positions have been reported to play a critical role in several functions of the NP protein. For example, the S314N mutation was shown to be responsible for the temperature-sensitive phenotype of A/WSN/33 [29]. Substitution of asparagine at position 319 to lysine (N319K) was found to augment viral replication, possibly through enhanced binding with importin α1 [31]. Whether changing V313 of NPS-OIV would have an effect on viral replication or virulence is currently being investigated.
In conclusion, our data collectively indicate that unique characteristics of NPS-OIV, including the early start codon and the V313 residue in NPS-OIV, are essential for the compatibility between NPS-OIV and the polymerase complex of S-OIV, which could potentially contribute to restriction of reassortment between S-OIV and the influenza virus used in this study, and possibly other human influenza viruses as well.
References
Faruqui F, Mukundan D (2009) Pandemic influenza: a review. Curr Opin Pediatr 22:530
Halasa NB (2010) Update on the 2009 pandemic influenza A H1N1 in children. Curr Opin Pediatr 22:83–87
Siston AM, Rasmussen SA, Honein MA, Fry AM, Seib K, Callaghan WM, Louie J, Doyle TJ, Crockett M, Lynfield R, Moore Z, Wiedeman C, Anand M, Tabony L, Nielsen CF, Waller K, Page S, Thompson JM, Avery C, Springs CB, Jones T, Williams JL, Newsome K, Finelli L, Jamieson DJ (2010) Pandemic 2009 influenza A(H1N1) virus illness among pregnant women in the United States. JAMA 303:1517–1525
Herfst S, Chutinimitkul S, Ye J, de Wit E, Munster VJ, Schrauwen EJ, Bestebroer TM, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Osterhaus AD, Perez DR, Fouchier RA (2010) Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J Virol 84:3752–3758
Jagger BW, Memoli MJ, Sheng ZM, Qi L, Hrabal RJ, Allen GL, Dugan VG, Wang R, Digard P, Kash JC, Taubenberger JK (2010) The PB2-E627K mutation attenuates viruses containing the 2009 H1N1 influenza pandemic polymerase. mBio 1:e00067-10
Zhu H, Wang J, Wang P, Song W, Zheng Z, Chen R, Guo K, Zhang T, Peiris JS, Chen H, Guan Y (2010) Substitution of lysine at 627 position in PB2 protein does not change virulence of the 2009 pandemic H1N1 virus in mice. Virology 401:1–5
Mehle A, Doudna JA (2009) Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci USA 106:21312–21316
Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O’Connor DH, Neumann G, Stewart LJ, Kawaoka Y (2010) Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog 6:e1001034
Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS, Guan Y (2010) Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529
Chen GW, Shih SR (2009) Genomic signatures of influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis 15:1897–1903
Pan C, Cheung B, Tan S, Li C, Li L, Liu S, Jiang S Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One 5:e9549
Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG (2000) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA 97:6108–6113
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289
Wanitchang A, Jengarn J, Jongkaewwattana A (2011) The N terminus of PA polymerase of swine-origin influenza virus H1N1 determines its compatibility with PB2 and PB1 subunits through a strain-specific amino acid serine 186. Virus Res 155:325–333
Yongkiettrakul S, Boonyapakron K, Jongkaewwattana A, Wanitchang A, Leartsakulpanich U, Chitnumsub P, Eurwilaichitr L, Yuthavong Y (2009) Avian influenza A/H5N1 neuraminidase expressed in yeast with a functional head domain. J Virol Methods 156:44–51
Kozak M (1992) Regulation of translation in eukaryotic systems. Annu Rev Cell Biol 8:197–225
Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P (2009) A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83:8021–8031
Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y (2007) Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81:30–41
Wu WW, Sun YH, Pante N (2007) Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol J 4:49
Belshe RB (2005) The origins of pandemic influenza—lessons from the 1918 virus. N Engl J Med 353:2209–2211
Schnitzler SU, Schnitzler P (2009) An update on swine-origin influenza virus A/H1N1: a review. Virus Genes 39:279–292
Li C, Hatta M, Nidom CA, Muramoto Y, Watanabe S, Neumann G, Kawaoka Y (2010) Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proc Natl Acad Sci USA 107:4687–4692
Li C, Hatta M, Watanabe S, Neumann G, Kawaoka Y (2008) Compatibility among polymerase subunit proteins is a restricting factor in reassortment between equine H7N7 and human H3N2 influenza viruses. J Virol 82:11880–11888
Octaviani CP, Ozawa M, Yamada S, Goto H, Kawaoka Y (2010) High genetic compatibility between swine-origin H1N1 and highly pathogenic avian H5N1 influenza viruses. J Virol 84:10918–10922
Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S (2008) Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol 62:403–424
Ye Q, Krug RM, Tao YJ (2006) The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078–1082
Biswas SK, Boutz PL, Nayak DP (1998) Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol 72:5493–5501
Medcalf L, Poole E, Elton D, Digard P (1999) Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J Virol 73:7349–7356
Poole E, Elton D, Medcalf L, Digard P (2004) Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321:120–133
Gabriel G, Herwig A, Klenk HD (2008) Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 4:e11
Acknowledgments
We are grateful to Drs. R.G. Webster and E. Hoffmann (St. Jude Children’s Research Hospital) for providing the plasmids for reverse genetics of the PR8 strain; and to Drs. Pathom Sawanpanyalert, Sathit Pichyangkul and Arunee Thititanyanont for providing viral samples. This work was supported in part by the National Science Development Agency (NSTDA) (CPMO-P-00-20386 grant).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wanitchang, A., Patarasirin, P., Jengarn, J. et al. Atypical characteristics of nucleoprotein of pandemic influenza virus H1N1 and their roles in reassortment restriction. Arch Virol 156, 1031–1040 (2011). https://doi.org/10.1007/s00705-011-0947-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-0947-6