Abstract
African swine fever virus (ASFV) encodes proteins that manipulate important host antiviral mechanisms. Bioinformatic analysis of the ASFV genome revealed ORF I329L, a gene without any previous functional characterization as a possible inhibitor of TLR signaling. We demonstrate that ORF I329L encodes a highly glycosylated protein expressed in the cell membrane and on its surface. I329L also inhibited dsRNA-stimulated activation of NFκB and IRF3, two key players in innate immunity. Consistent with this, expression of I329L protein also inhibited the activation of interferon-β and CCL5. Finally, overexpression of TRIF reversed I329L-mediated inhibition of both NFκB and IRF3 activation. Our results suggest that TRIF, a key MyD88-independent adaptor molecule, is a possible target of this viral host modulation gene. The demonstration of an ASFV host evasion molecule inhibiting TLR responses is consistent with the ability of this virus to infect vertebrate and invertebrate hosts, both of which deploy innate immunity controlled by conserved TLR systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
African swine fever virus (ASFV) is a lethal haemorrhagic pathogen of domestic pigs. In its natural mammalian hosts, the Warthog and Bushpig, however, the virus is persistent, non-pathogenic and transmitted by an invertebrate vector, the soft tick. The virus may therefore be considered as a veterinary model for recently emerged haemorrhagic infections.
AFSV is an enveloped DNA virus with icosahedral morphology that is now classified as the only member of a new virus family, Asfarviridae [1]. It exhibits a characteristic genomic structure, large numbers of novel genes and a cytoplasmic replication strategy, and, in contrast to all other DNA viruses, this arbovirus, infects both vertebrate (swine) and invertebrate (tick) hosts. The ability of the virus to infect macrophages and to persist in its natural hosts, and in domestic pigs, which recover from infection with less virulent isolates, shows that the virus has effective mechanisms to evade host defense systems, in particular, innate immunity. It is estimated that half to two-thirds of the approximately 150 genes encoded by ASFV do not have known viral or cellular homologs [2]. Most of the structural proteins and enzymes have been identified, but many of these non-homologous genes have no homology to known proteins or enzymes. Therefore, we hypothesize that some of these genes have evolved for, or may be involved in, host evasion. Such genes provide a source of potentially valuable tools for understanding virus pathogenesis and for identifying novel aspects of the vertebrate immune system.
The fact that ASFV has adapted to infect both mammalian macrophages and an invertebrate tick host suggests that this virus may have evolved immune evasion genes focused on innate immunity, more specifically, for manipulation of the Toll-like receptor (TLR) signaling system, which is conserved in both mammals and arthropods. Moreover, there is abundant expression of TLRs in macrophages, where they play a critical role in the detection of viral infections, leading to the inflammatory response and induction of an interferon (IFN)-mediated antiviral state in infected and neighboring cells [3]. Under some circumstances, however, uncontrolled activation of macrophages leads to undesirable pathological consequences.
To date, there are at least 13 distinct mammalian TLRs, all sharing similarities in their extracellular and intracellular domains, in particular, the conserved intracellular Toll/IL-1 receptor (TIR) domain [4, 5]. Dimerization of TLRs induced by interaction with the corresponding ligands initiates the intracellular signaling cascade, followed by recruitment of intracellular adaptor proteins to their dimerized intracellular domains [6]. Among the adaptor proteins, myeloid differentiation primary response gene (MyD88) and TIR-domain-containing adaptor-inducing interferon-β (TRIF) are critical for signaling, and hence, TLR signaling falls into the MyD88-dependent and TRIF-dependent pathways [7, 8], both terminating in activation of the transcription factors nuclear factor κB (NFκB) and activator protein-1 (AP-1).
Binding to each TLR induces common effects, such as induction of inflammatory cytokines and chemokines and more specific and restricted activities, such as the induction of interferon-β (IFN-β). In turn, IFN-β amplifies the IFN response and the development of antiviral activity, thus contributing to an important defense against viral infections [9]. The transcription factors interferon regulatory factor-3 (IRF-3) and interferon regulatory factor-7 (IRF-7) are master regulators of type I IFN activation and participate in both the TLR-dependent and -independent pathways of innate immune responses to viral pathogens [10]. Type I IFNs not only induce an antiviral state in most cells but also have diverse functions in the development of adaptive immunity. The existence of several mechanisms in the immunocompetent host to avoid infections by pathogens has provided the selective pressure for successful viral pathogens to acquire strategies during evolution to evade the host immune responses [11], in particular for inhibiting and regulating TLR function, with several targeting the MyD88-dependent pathway [12].
Viral infections are frequently sensed by TLR3, which detects dsRNA produced during replication of both DNA and RNA viruses. MyD88-independent signaling through the TLR3 pathway is initiated by the recruitment of the adaptor TRIF, leading to the subsequent production of inflammatory cytokines and type I IFNs and the up-regulation of co-stimulatory molecules. The importance of TLR3 signaling is emphasized by the fact that it is a target for immune evasion by viruses [13], for example, vaccinia virus A52R [14–16] and A46R [14, 17] proteins or hepatitis C virus (HCV) NS3/4A protease [18, 19]. Downstream of TRIF, Sterile-alpha and Armadillo motif-containing protein (SARM), another cellular molecule with a TIR domain, provides an important regulatory point in the MyD88-independent pathways [20]. Viruses may benefit from disrupting TLR signaling, and indeed it has been suggested that vaccinia virus directly engages TLRs [21, 22]. Taking into account the complexity of pathogen recognition receptor (PRR) signaling in the innate immune response, it is likely that viruses have evolved a whole range of mechanisms targeting different PRRs and at different levels of their signaling pathways. Thus, the study of virus evasion strategies is not only directly relevant to understanding virus pathogenesis but can also define novel aspects of our own immune system. For example, in HCV infection, the viral NS3/4, a protease complex targets and cleaves the interferon promoter stimulator-1 (IPS-1) adaptor protein to ablate signaling of immune defenses controlled by IFN α/β. Similarly, the finding that cleavage of IPS-1 by NS3/4 releases it from the mitochondrial membrane revealed that the mitochondrial attachment of IPS-1 is functionally important [19].
Here, we report a successful search for an ASFV host evasion gene that inhibits an important component of the innate immune response controlled by a TLR. A combination of bioinformatics, biochemical and reporter gene assays identified the ORF I329L, a late-expressed protein [23], which was characterized as a type I transmembrane protein with homology to TRIF in the C-terminal region and shown to be capable of inhibiting the TLR3 signaling pathway through an as yet undefined effect on the critical intracellular signaling adaptor molecule TRIF.
Materials and methods
Bioinformatic analysis
Full-length sequences of non-assigned ASFV ORFs were screened for homologies using protein-protein BLAST from the National Center for Biotechnology Information (NCBI). Screening for patterns or domains was performed using InterProScan 17.0 from the European Molecular Biology Laboratory (EMBL) and Prosite 20.33 databases. The transmembrane region prediction was performed by TransMembrane Helix prediction using the Hidden Markov Models 2.0 (TMHMM) program [24], and 3D-structure similarities were analyzed with Protein Homology/analogy Recognition Engine (Phyre) [25]. Multiple sequence alignments were made with ClustalW2 from the European Bioinformatics Institute (EBI) website.
Plasmids
Plasmids pcDNA3-HA and pHR-CMV-eGFP were gifts from Dr. L. Dixon (Institute for Animal Health, Pirbright, UK) and Dr. Y. Ikeda (University College London, London, UK), respectively. The pFlag-TRIF- and pCCL5-promoter-containing plasmids were gifts from Dr. Andrew G. Bowie (Trinity College, Ireland), pDsRed-Monomer-Golgi and pDsRed2-ER vectors were purchased from ClonTech (catalogue number 632480 and 632409, respectively). The β-gal-CMV- (β-gal), the NFκB- (pPRD2) and IRF3- and IFN-β-promoter-containing plasmids were gifts from Dr. S. Goodbourn (St George’s University, London). The plasmids expressing TLR3 and CD4-TLRs chimaeras were provided by Dr. R. Medhzitov (Yale University, USA).
Cloning of I329L
The ASFV ORF I329L (accession number NP_042833) was obtained by PCR amplification from DNA of the tissue-culture-adapted non-pathogenic ASFV isolate Ba71V (Uniprot database, code I329_ASFB7) with primers incorporating restriction sites for BamHI upstream and EcoRV downstream of the ORF and using the high-fidelity enzyme Pfu DNA polymerase. The primers used were 5’caggatccatgctaagggttttcatattttttg3’, 5’ctgatatcctttcttcttgaacatgaaacc3’. The amplified product was identified with ethidium bromide visualization on an agarose gel based on the expected size of the amplicon, and the DNA was purified from the excised band. The fragment was then cloned into plasmid pcDNA3-HA, fused in frame with an N-terminal influenza haemagglutinin (HA) peptide tag. The fidelity of the sequence of the cloned fragment was confirmed by automated sequencing.
For construction of a recombinant lentivirus vector (pHR-CMV-HA-I329L-eGFP), the I329L gene was excised from pcDNA3, together with the HA tag, and cloned into the vector pHR-CMV-eGFP upstream of an internal ribosome entry site (IRES)-driven enhanced green fluorescent protein gene (eGFP).
Cells
Human HEK-293T and murine NIH-3T3 cell lines were cultured in 5% CO2 at 37°C in Dulbecco’s modified Eagle medium (DMEM) with GlutaMAXTM-I and 1000 mg/l glucose, supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS) and 100 U/ml and 100 μg/ml penicillin and streptomycin, respectively (all products from Invitrogen). The stable cell line 293-hTLR3-HA (HEK-TLR3), purchased from InvivoGen®, was maintained under the same conditions and cultured in regular DMEM supplemented with 10 μg/ml blasticidin [26].
Lentivirus production
Lentivirus was produced by transient transfection of HEK-293T cells with a weight ratio of 3:1:1 of vector to packaging to envelope plasmids, respectively, using FuGENE 6 (Roche) according to the manufacturer’s instructions. Control lentivirus was produced by cotransfection of the packaging and envelope plasmid together with the empty pHR-CMV-eGFP plasmid. For production of recombinant lentivirus expressing I329L, the plasmid pHR-CMV-HA-I329L-eGFP was used. Supernatants containing the lentivirus were collected at 48 h and 72 h post-transfection and clarified by centrifugation, and the lentivirus was collected by ultracentrifugation (125,000 x g, 3 h, 4°C). Virus pellets were resuspended in fresh culture medium, frozen at −80°C and titrated by infection of HEK-293T cells with a dilution factor of 4. Analysis of lentivirus-infected cells was done by detecting eGFP-positive cells by flow cytometry at 48 h postinfection (p.i.).
Lentivirus transduction of NIH-3T3 cells with I329L
The NIH-I329L and NIH-eGFP stable cell lines were produced by lentivirus infection of NIH-3T3 cells with the recombinant pHR-CMV-HA-I329L-eGFP and empty pHR-CMV-eGFP (prepared as described in the section “Cloning of I329L”), respectively, using a multiplicity of infection (m.o.i.) of 10 in DMEM. Analysis of NIH-I329L to determine the percentage of transduced cells was done by detecting eGFP-positive cells by flow cytometry at 48 h post-infection (p.i.), and this was verified to be greater than 99%.
Antibodies
The antibodies used in this study were as follows: rat monoclonal high-affinity antibody (clone 3F10) against HA conjugated with horseradish peroxidase (HRP) (Roche, 12013819001), rabbit polyclonal affinity-purified antibody against HA for immunoprecipitation assays (Sigma H6908), rat monoclonal high-affinity antibody (clone 3F10) against HA for immunofluorescence assays (Roche, 11867423001), donkey affinity-purified anti-rat IgG antibody conjugated with AMCA (Jakson Immuno Research, 712-155-150), mouse monoclonal antibody (clone B-5-1-2) against α-tubulin (Sigma-Aldrich, T6074) and rabbit polyclonal anti-mouse Immunoglobulins conjugated with HRP (Dako, P0161).
Immunoblotting
NIH-I329L cells were cultured in 75-cm2 flasks at 37°C, under 5% CO2, in DMEM with GlutaMAXTM-I and 1000 mg/l glucose, supplemented with 10% (v/v) heat-inactivated FCS and 100 U/ml and 100 μg/ml penicillin and streptomycin, respectively (all products from Invitrogen). Cells were lysed in cold lysis buffer (6 M urea, 75 mM NaCl, 1 mM EDTA, 1% (v/v) NP40, 2% (v/v) glycerol, 1 mM PMSF, 1 μM DTT, 25 mM HEPES, pH 7.4). After protein quantification by the Bradford method using the Bio-Rad Protein Assay reagent, extracts were boiled (5 min, 100°C) with sample buffer (1.7% [w/v] SDS, 5% [v/v] glycerol, 0.1 M DTT, bromophenol blue [0.02 mg/ml], 58 mM Tris-HCl, pH 6.8). Proteins were then separated by 10-12% SDS-PAGE and electroblotted onto polyvinylidene fluoride (PVDF, BioRad) membranes and blocked with PBS-5% (w/v) non-fat dried milk for 1 h at room temperature (RT). The development of the blots to reveal recombinant viral proteins was performed with a rat monoclonal high affinity antibody against HA conjugated with HRP (Roche) by incubating for 1 h at RT or overnight at 4°C, and the identified molecules were revealed with ECL chemiluminescent reagents (PerBio Science) according to the manufacturer’s instructions. Blots were also developed with a mouse monoclonal antibody against α-tubulin (Sigma-Aldrich), followed by rabbit polyclonal anti-mouse immunoglobulins-HRP (Dako) to provide an internal control for protein loading.
Immunoprecipitation
Mouse NIH-I329L and mouse NIH-eGFP cells were collected by scraping from 75-cm2 flasks and lysed using cold RIPA buffer (6 M Urea, 75 mM NaCl, 1 mM EDTA, 1% (v/v) NP40, 2% (v/v) glycerol, 1 mM PMSF, 5.5 μg/ml protease inhibitors (SIGMA) and 1 μM DTT, 25 mM HEPES, pH 7.4) (1 ml per 7 × 106 cells). The resulting protein extract was centrifuged at 20,000 × g for 10 min at 4°C to remove cellular debris. The supernatant of the protein extracts was collected and, after determining the concentration of protein, was pre-cleared by adding 100 μl of protein G beads. After rotating for 1 h at 4°C, the beads were removed by centrifugation, and the recombinant I329L was recovered from the supernatant by adding protein G beads (100 μl) and 5 μg of rabbit polyclonal affinity-purified anti-HA antibody (Sigma). Following rotation for 1.5 h at 4°C, the beads were then washed twice with lysis buffer without NP-40 and urea, and finally heated at 100°C for 10 min with loading buffer (2% [w/v] SDS, 2% [v/v] β-mercaptoethanol, 0.1% [w/v] bromophenol blue, 10% [v/v] glycerol, 50 mM Tris-HCl, pH 6.8) for 10 min, and centrifuged for 30 sec. The supernatants were then loaded onto an SDS-PAGE gel (10%).
Cell-surface biotinylation
Cell-surface biotinylation was done using a kit purchased from Pierce (#21335). One 75-cm2 flask with transduced NIH-I329L cells was cultured until the cells reached 95% confluence. Then, the cells were washed twice with cold PBS and EZ-Link Sulfo-NHS-LC-Biotin dissolved in cold PBS (0.5 mg/ml) was added to the cells, followed by incubation on a rocking platform with gentle agitation (30 min, 4°C). To stop the reaction, 500 μl of a “quenching” solution (supplied by the manufacturer without information) was added to each flask. Cells were then scraped off, centrifuged, washed with TBS and lysed in modified RIPA buffer (1% [v/v] Triton X-100, 0.2% [w/v] Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 0.2% (w/v) SDS, 1 mM PMSF, protease inhibitors, 50 mM Tris-HCl, pH 7.4). The biotinylated surface proteins were recovered from the detergent lysate with streptavidin beads (Pierce Kit, #89881) (overnight, 4°C, in an orbital shaker). The streptavidin beads were centrifuged, washed 3 times with lysis buffer without detergent, and finally suspended in 100 μl 1× loading buffer (2% [w/v] SDS, 2% [v/v] β-mercaptoethanol, 0.1% [w/v] bromophenol blue, 10% [v/v] glycerol, 50 mM Tris-HCl, pH 6.8). The samples were heated (10 min, 100°C) and centrifuged, and the supernatants were loaded onto 10% SDS-PAGE gel. After electrophoresis, the proteins were transferred to a PVDF membrane (Bio-Rad) and blocked with 5% non-fat dried milk (1 h, RT). The biotinylated HA-I329L protein was revealed with a rat monoclonal high-affinity antibody against HA conjugated with HRP (Roche), followed by ECL chemiluminescent reagents (PerBio Science) according to the manufacturer’s instructions.
Deglycosylation assay
Lentivirus-transduced NIH-I329L cells were collected, and the recombinant I329L was immunoprecipitated as described above. Bound I329L was eluted from the anti-HA protein G beads by heating (10 min, 100°C) in 0.5% (w/v) SDS with 10% (v/v) β-mercaptoethanol. For deglycosylation, the eluted sample was treated with peptide-N-glycosidase F (PNGaseF) (Roche) or endoglycosidase H (Endo H) (New England Biolabs) according to the manufacturer’s instructions. As controls, the enzymes were omitted. Proteins were then precipitated with ethanol at 80% (v/v) (overnight, −20°C). The pellet was resuspended in 20 μl of SDS-PAGE loading buffer, heated (10 min, 100°C), and loaded onto an SDS-PAGE gel.
Immunofluorescence to demonstrate co-localization of I329L with ER and Golgi
Mouse NIH-I329L cells were seeded onto coverslips (pretreated with 1% [v/v] poly-L-lysine) and incubated overnight at 37°C and 5% CO2. For double staining to reveal the endoplasmatic reticulum and the trans-medial region of the Golgi apparatus, the cells were transfected using Lipofectamine 2000 (Invitrogen) with 2 μg of a plasmid coding for the ER retention sequence KDEL (plasmid pDsRed2-ER, Clontech) or coding for a sequence corresponding to the N-terminal 81 amino acids of human beta-1,4-galactosyltransferase (GT) (plasmid pDsRed-Monomer-Golgi, Clontech) fused with the fluorescent red protein.
After transfection for 24 h, the cells were fixed with 4% (w/v) paraformaldehyde in PBS for 20 min at RT and then incubated in a blocking solution (PBS containing 0.05% Tween-20 and 5% normal goat serum) (30 min, RT). The cells were permeabilized using PBS containing 0.1% (v/v) Triton X-100. To detect recombinant I329L, a primary rat monoclonal high-affinity antibody against HA diluted in blocking solution was used (1 h, RT), followed by a donkey affinity-purified anti-rat IgG antibody conjugated with AMCA (1 h, RT). All washes after incubation with antibody were performed at RT with PBS containing 0.05% (v/v) Tween-20. Cells were mounted in Vectashield (Vector Laboratories) and observed under a fluorescence microscope. HA-I329L is shown with a green colour for better visual perception. Fluorescent images were taken with a Leica DMRA2 microscope equipped with a cooled CCD camera (Leica. Corp., Wetzlar, Germany).
Luciferase reporter assays
For the CD4-TLR reporter gene assay, HEK-293T cells were cultured in 24-well plates at 6x104/well, and 24 h later, they were transfected using Lipofectamine 2000 (Invitrogen) with three plasmids. One, the NFκB-driven luciferase reporter contained only the positive regulatory domain, PRD2, of the NFκB binding site of the IFN-β promoter (NFκB) (100 ng), the second contained β-gal-CMV (β-gal) (25 ng), and the third chimeric plasmid contained a CD4 extracellular domain co-ligated upstream of a TLR1 (CD4-TLR1) or TLR3 (CD4-TLR3) intracellular domain (50 ng). The effect of the co-transfection of the I329L gene cloned into the pcDNA3 plasmid (pcDNA3-I329L) (300 ng) provides an assay for interference of the viral protein with the TLR signaling pathway [27]. Empty pcDNA3 served as the negative control (pcDNA3). After incubating for 48 h at 37°C, the cells were lysed in 100 μl lysis solution (ABX210LM, Promega Systems) according to the manufacturer’s instructions, and samples were assayed for both luciferase and β-galactosidase activities. The luciferase activity was normalized to the β-galactosidase activity from the co-transfected plasmid internal control and expressed as luciferase relative to galactosidase activity. Values are expressed as mean relative stimulation ± SD (calculated from triplicate determinations). A minimum of three independent assays were done for each experiment reported.
In similar reporter assays, HEK-TLR3 cells were transfected with reporter plasmids for IFN-β, NFκB, CCL5, and the internal control β-gal, and expression plasmids pcDNA3-I329L or pcDNA3 (negative control), keeping the total amounts of DNA and equimolar ratios constant in all assays by adding the appropriate amount of empty vector. After 48 h, cells were stimulated with polyinosinic:polycytidylic acid (poly (I:C)) or IL1-β (PeproTech) or TNF-α (PeproTech) as indicated in the figures for 5 hours. The cells were lysed and assayed for luciferase as described above.
IFN-β reporter assays were also done using HEK-293T cells ectopically expressing TRIF. Transfections were performed with equal amounts of DNA mixtures comprising 25 ng of TRIF plasmid vector (TRIF), the I329L plasmid vector (pcDNA3-I329L) (0, 100, 200 and 300 ng), the IFN-β-reporter plasmids (IFN-β) (100 ng) and the β-gal internal control plasmid (β-gal) (25 ng). The pcDNA3 negative control was included in the transfection whenever necessary in order to keep the total amounts of DNA and equimolar ratios constant in all assays. After 48 h, the cells were stimulated for 6 hours with poly (I:C) (25 μg/ml) where indicated, before harvesting the cells. Lysis and luciferase activity were measured and normalized to the β-galactosidase activity as described above.
Finally, to determine the impact of intracellular stimulation with poly (I:C), HEK-293T cells were transfected with 300 ng empty vector (pcDNA3) or 300 ng I329L plasmid vector (pcDNA3-I329L) together with 100 ng of IFN-β plasmid (IFN-β). After 48 h, the cells were stimulated for 6 h with intracellularly delivered poly (I:C) (0.5 μg/ml) using Lipofectamine 2000 (Invitrogen). Lysis and luciferase activity were measured and normalized to the β-galactosidase activity as described above.
Statistical analysis
Data are expressed as mean ± SD and were analyzed for significance using Student’s T test. P-values are as follows: *** P < 0.005, ** P < 0.01 and * P < 0.05.
Results
Identification of ASFV ORF I329L as a potential homologue of the TLR family
Bioinformatic analysis of the sequence of pI329L from the tissue-culture-adapted non-pathogenic ASFV isolate Ba71V (Uniprot database code I329_ASFB7), which grows in a variety of commonly used fibroblast cell lines, reveals a type I membrane structure with a 17-amino-acid signal peptide (amino acids 1-17), followed by an N-terminal extracellular domain (amino acids 18-239), a transmembrane domain (amino acids 240-260), and a 69-amino-acid C-terminal intracellular domain (amino acids 261-329). A number of potential glycosylation sites were indicated, at amino acids 32, 39, 44, 76, 82, 101, 185, and 219. Simple sequence examination of the sequence database did not show any gene similar to I329L; however, after multiple alignments with several TLR proteins, a weak but interesting alignment between I329L and TLR from Drosophila melanogaster was revealed. Importantly, the intracellular domain of I329L aligned with BOX1 and BOX2 regions of the human TIR-like domain of TLR3 (Fig. 1A), with a sequence similarity of 35% to TLR3 in BOX1 (Fig. 1B). Furthermore, the BOX2 of TLR3 also showed similarities with the I329L intracellular domain (Fig. 1C). Finally, the conserved extracellular region with leucine-rich repeats (LRRs) is an important motif for interaction between proteins and is common to several TLRs. Although the observed similarity was admittedly weak, we nonetheless decided to clone and test I329L for its impact on poly (I:C)-stimulated innate immunity in reporter assays (see below).
Homology with TLR3 in the intracellular domain of ASFV protein I329L. The analysis presented here was performed with ClustalW2. 1A) Sequence of the intracellular domain of I329L (amino acid residues 260 to 329) indicating the predicted BOX1 and BOX2 regions. 1B) Predicted TIR-like domain in the BOX1 domain of I329L has sequence homology with known proteins containing the BOX1 TIR domain. 1C) Predicted TIR-like domain in the BOX 2 domain of I329L. Details are in Materials and methods
The cellular distribution of ASFV ORF I329L is consistent with ER, Golgi and plasma membrane localization
Bioinformatic analysis and demonstration of extensive glycosylation suggested that the I329L protein was a typical transmembrane protein. This possibility was confirmed in I329L-lentivirus-transduced NIH-3T3 cells (NIH-I329L) by immunofluorescence and cell-surface labelling. For immunofluorescence of I329L, the I329L protein was detected with a rat monoclonal high-affinity antibody (clone 3F10) against HA, followed by a donkey affinity-purified anti-rat IgG antibody conjugated with AMCA. Co-detection of the endoplasmic reticulum (ER) and Golgi apparatus was achieved by simultaneously transfecting the cells with two marker plasmids: one coding for red-fluorescent protein (dsRed) fused with the KDEL sequence in order to target ER (plasmid pDsRed2-ER) and the other coding for dsRed fused with an integral protein of the Golgi apparatus (plasmid pDsRed-Monomer-Golgi). As can be seen, the cellular distribution of I329L corresponds to that of the markers for ER and Golgi (Fig. 2A and B), indicating that I329L passes through the secretory pathway. Finally, since the intracellular secretory pathway frequently culminates the incorporation of glycoproteins into the cell membrane, viable NIH-I329L-transduced cells were surface-biotinylated. Subsequent immunoprecipitation with streptavidin and western blotting with anti-HA to detect I329L confirmed the surface expression of I329L in the transduced cells (Fig. 2D).
ASFV protein I329L is highly glycosylated and localizes to cell membranes. 2A) I329L protein distribution is consistent with endoplasmic reticulum localization. Lentivirus-transduced NIH-I329L cells were transfected with plasmid pDsRed2-ER to localize the protein to the ER (red, left) and then co-stained with a rat monoclonal high-affinity antibody against the HA, followed by a donkey affinity-purified anti-rat IgG antibody conjugated with AMCA (left, middle). The colocalization is evident in the merged image (right panel). 2B) I329L protein distribution is consistent with Golgi localization. The NIH-I329L cells were transfected with plasmid pDsRed-Monomer-Golgi to localize the protein to the Golgi (red, left) and then co-stained with a rat monoclonal high-affinity antibody against the HA, followed by a donkey affinity purified anti-rat IgG antibody conjugated with AMCA (left, middle). The colocalization is evident in the merged image (right panel). 2C) The I329L protein was immunoprecipitated from lysates from the lentivirus-transduced NIH-I329L cell line with rabbit anti-HA antibodies bound to protein G beads. The bound protein was eluted and digested with EndoH (a) or PNGase (c) and examined by SDS-PAGE. Controls samples were treated with only EndoH buffer (b) or only PNGase buffer (d). 2D) I329L protein is expressed at the cell surface. Biotinylated surface proteins from biotynylated NIH-I329L cells were collected with streptavidin beads, loaded onto a 10% SDS-PAGE gel and transferred to PVDF membranes, and I329L was revealed with a rat monoclonal antibody against HA conjugated with HRP (band in lane NIHI329L on the left). Lane NIHeGFP represents NIH-3T3 cells transduced with the empty lentivirus. Details are in Materials and methods
ASFV ORF I329L codes for a highly glycosylated protein
Expression of HA-I329L in NIH-3T3 cells was accomplished by transduction with HA-I329L recombinant lentivirus (NIH-I329L). The protein immunoprecipitated with anti-HA antibody migrated in SDS-PAGE with an apparent molecular weight of ~50 kDa, significantly larger than the molecular weight predicted from the amino acid sequence alone (Fig. 2C, lanes b and d), thus raising the possibility of extensive glycosylation, as indeed is suggested by bioinformatic predictions. Therefore, the recombinant I329L protein was immunoprecipitated from NIH-I329L cells and digested with two endoglycosidases, PNGase F, which digests high-mannose, hybrid and complex types of oligosaccharides, and endoglycosidase H (EndoH), which digests only high-mannose and hybrid types of oligosaccharides. Digestion with EndoH and PNGase F yielded recombinant proteins that migrated with a molecular mass of ~37 kDa (Fig. 2C, lane a), and ~36 kDa (Fig. 2C, lane c), respectively, thus confirming that the I329L protein is extensively glycosylated. Furthermore, the molecular weight observed for the sample after digestion with PNGase F was lower than after digestion with Endo H, suggesting that I329L has a small number of glycans of the complex type.
ASFV ORF I329L inhibits TLR3 signaling
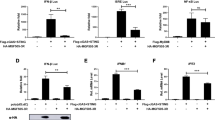
An important sensor of viral infections is TLR3, which detects dsRNA and signals through a MyD88-independent pathway. In order to demonstrate that I329L was interfering with TLR3 signaling, we used two entirely different experiments. In the first approach, we used a CD4-TLR assay focusing on an NFκB-driven luciferase reporter. These CD4-TLR constructs comprise the murine CD4 extracellular domain co-ligated to the cytoplasmatic domain of a human TLR. The extracellular CD4 domain dimerizes spontaneously, resulting in dimerization of the intracellular TLR domain and thus constitutive activation of NFκB, which is detected using the simultaneously transfected luciferase reporter plasmid. Using this system, expression of I329L was found to inhibit the induction of the NFκB-dependent reporter gene signal (Fig. 3A). Negative controls consisted of plasmid pcDNA3 coding for H171R, a known structural viral protein of ASFV, instead of the I329L gene, and a CD4-TLR1 construct instead of CD4-TLR3 (Fig. 3A).
I329L inhibits NFκB activation by TLR3.3A) I329L inhibits activation of NFκB. HEK-293T cells were transfected with constitutively active CD4-TLR plasmids (50 ng CD4-TLR1 or CD4-TLR3) in the presence of 300 ng of I329L plasmid (pcDNA3-I329L), 100 ng of NFκB reporter plasmid (NFκB) and 25 ng of β-gal reporter plasmid (β-gal) (black bars) and incubated for 48 h prior to reporter assays. The negative controls were the plasmid vector without insert (pcDNA3) (white bars) or with the control H171R insert (pcDNA3-H171R) (grey bars). 3B) I329L inhibits poly (I:C)-dependent activation of NFκB. HEK-293T cells were co-transfected with plasmid vectors encoding 50 ng TLR3 (TLR3), 100 ng NFκB reporter plasmid (NFκB), 300 ng ASFV I329L gene (pcDNA3-I329L) and 25 ng of β-gal reporter plasmid (β-gal) (black bars). Negative and positive controls were the plasmid vector of I329L gene without an insert (pcDNA3) and TLR3 (grey bars), and the TLR3 plasmid alone (white bars). The cells were stimulated with poly (I:C) at the concentration indicated. Luciferase activity was normalized to the β-galactosidase activity obtained with the co-transfected β-gal plasmid internal control. Standard deviations are shown by error bars. Details are in Materials and methods
As the synthetic dsRNA analogue polyinosine-polycytidylic acid (poly (I:C)) activates cells via TLR3, in the second system, we also tested the effect of I329L on TLR3-dependent NFκB activation induced by poly (I:C). Cells (HEK-293T) were co-transfected with a construct encoding TLR3, the luciferase reporter containing the NFκB (PRD2) binding sequence, and the plasmid containing the cloned ASFV I329L gene. The cells were stimulated with increasing concentrations of poly (I:C) (0, 5, 25, and 50 μg/ml). The vector of the I329L gene without an insert (pcDNA3) was used as negative control. This TLR3-dependent activation of NFκB in HEK-293T cells expressing TLR3 was inhibited by the ASFV gene I329L at all concentrations of the ligand, poly (I:C), that were tested (Fig. 3B). Thus, I329L was shown by two independent approaches to inhibit the TLR3-mediated activation of NFκB.
The I329L gene mediates MyD88-independent induction of poly (I:C)-stimulated activation of NFκB and IRF3
To confirm and further explore the mechanisms of I329L-mediated inhibition of TLR3 activation, human embryonic kidney fibroblasts (HEK-293T) stably expressing TLR3 (HEK-TLR3) were transfected together with an NFκB luciferase reporter plasmid (NFκB) and stimulated with poly (I:C) or IL-1β. As can be seen, the poly (I:C)-stimulated activation of NFκB via the established TLR3 pathway was inhibited by expression of I329L (Fig. 4A). Activation of NFκB can occur either dependently or independently of the MyD88 adaptor molecule. However, the absence of an effect on the IL-1 receptor pathway excludes the possibility that I329L modulates TLR signaling by interacting with the adaptor molecule MyD88. Finally, we failed to find any inhibitory effect of I329L on NFκB activation stimulated by TNF-α, indicating that I329L does not have impact on the NFκB activation pathway via molecules other than those downstream of TLR ligation. Taking these results together suggests that I329L acts upstream of the IκB kinase (IKK-α/IKK-β/IKK-γ) complex, which in the case of TLR3 signalling could be TRIF, a possibility explored below.
I329L inhibits NFκB and IRF3 activation through an MyD88-independent pathway. 4A) I329L inhibits activation of NFκB via Poly I:C but not TNF-α or IL-1. HEK-TLR3 cells were co-transfected with 300 ng of empty plasmid vector (pcDNA3) or 300 ng of the same plasmid vector encoding I329L (pcDNA3-I329L), together with 100 ng of the NFκB luciferase reporter plasmid (NFκB) and 25 ng of β-galactosidase reporter plasmid (β-gal), and cultured for 48 h. At 6 h before harvesting, cells were not stimulated (white bars), or stimulated with IL-1β (100 ng/ml) (grey bars), TNF-α (100 ng/ml) (hatched bars) or poly (I:C) (25 μg/ml) (black bars). The NFκB reporter gene activity was then measured. 4B) I329L inhibits activation of IRF3. HEK-TLR3 cells were transfected with 300 ng empty vector (pcDNA3) or 300 ng of the same plasmid encoding I329L (pcDNA3-I329L) together with 100 ng of IRF3 (IRF3) and 25 ng of β-galactosidase reporter plasmids (β-gal), and 6 h before harvesting, cells were stimulated extracellularly with 25 μg/ml poly (I:C). Luciferase activity was normalized to the β-galactosidase activity given by the co-transfected β-gal plasmid internal control. Standard deviations are shown by error bars. Details are in Materials and methods
Signal transduction initiated through TLR3 ultimately diverges into two signaling branches, leading to activation of NFκB and IRF3. In order to determine whether I329L also inhibits activation of IRF3, HEK-TLR3 cells were co-transfected with I329L (pcDNA3) and a reporter plasmid containing the promoter of IRF3 (IRF3) and then stimulated with poly (I:C). As can be seen (Fig. 4B), in the presence of I329L, activation of the IRF3 promoter was significantly reduced.
I329L ASFV ORF inhibition of activation of IRF3 and subsequent expression of IFN-β and CCL5 responses
As TLR3 activates and programs interferon and chemokine expression through the coordinated activation of IRF3 and NFκB [28, 29], we next examined the effect of I329L on the activation of CCL5 expression using an appropriate reporter plasmid. We also extended the previously presented I329L-mediated inhibition of the NFκB-restricted PRD2 by using a reporter plasmid containing the entire IFN-β sequence. The rationale for this was that inhibition of activation of NFκB and IRF3 is predicted to inhibit induction of IFN-β and chemokine responses, two major effectors of the antiviral response induced by TLR3.
As can be seen, poly (I:C)-induced CCL5 activation from HEK-293T cells stably expressing TLR3 (HEK-TLR3) was inhibited by I329L (Fig. 5A). Moreover, poly (I:C)-induced IFN-β activation was also inhibited by I329L in HEK-TLR3 cells (Fig. 5B). Both results are consistent with an I329L-mediated inhibition of an MyD88-independent activation pathway [30] and raise the possibility that I329L targets TRIF, as TRIF is the only known adaptor protein that interacts with TLR3 and mediates induction of IFN-β through activation of NFκB and IRF3.
I329L inhibits activation of CCL5 and IFN-β promoter. 5A) I329L inhibits activation of CCL5. HEK-TLR3 cells were transfected with 300 ng empty plasmid vector (pcDNA3) or 300 ng I329L plasmid vector (pcDNA3-I329L) together with 100 ng of CCL5 reporter plasmid vector (CCL5). Six h before harvesting, cells were stimulated extracellularly with 25 μg/ml poly (I:C). 5B) I329L inhibits activation of IFN-β. HEK-TLR3 cells were transfected with 300 ng empty vector (pcDNA3) or 300 ng of I329L plasmid vector (pcDNA3-I329L) together with 100 ng of IFN-β plasmid vector (IFN-β). Six h before harvesting, cells were stimulated extracellularly with 25 μg/ml poly (I:C). 5C) I329L does not inhibit intracellular IFN-β activation independently of TLR3. HEK-293T cells were transfected with 300 ng empty vector (pcDNA3) or 300 ng I329L plasmid vector (pcDNA3-I329L) together with 100 ng of IFN-β plasmid reporter (IFN-β). Six h before harvesting, cells were stimulated intracellularly using Lipofectamine 2000 with 0.5 μg/ml poly (I:C). Luciferase activity was normalized to the β-galactosidase activity obtained with the cotransfected β-gal plasmid internal control. Standard deviations are shown by error bars. Details are in Materials and methods
The CCL5 promoter is under the control of the PRD2 and ISRE domains (binding NFκB and IRF3, respectively), the cAMP responsive element (CRE), AP-1, the gamma interferon activation site (GAS), the nuclear factor of activated T-cells (NFAT) and CCAAT/enhancer-binding protein (C/EBP) domains (binding proteins c-Jun/activating transcription factor 2 (ATF2), Fos/c-Jun, signal transducer and activator of transcription (STAT), NFAT and C/EBPβ. In our experimental system, the induction of CCL5 and IFN-β is the consequence of a poly (I:C)-stimulated MyD88-independent activation of TLR3. Thus, the possibility of an MyD-88-dependent, IRF5-mediated activation of CCL5 transcription can be excluded.
The possibility that I329L inhibits IFN-β activation induced intracellularly via a receptor other than TLR3 was investigated by introduction of poly (I:C) into HEK-TLR3 cells. Interestingly, there was no effect of I329L on IFN-β activation via an intracellular poly (I:C) stimulus (Lipofectamine-mediated poly (I:C) transfection) (Fig. 5C), and we therefore exclude the hypothesis that the effect of I329L is mediated via intracellular receptors such as melanoma differentiation associated gene 5 (MDA-5) or retinoic-acid-inducible protein I (RIG-1).
Taken together, these results suggest that I329L inhibits induction of promoters that are known to contain NFκB or IRF3 binding sites via the TLR3 pathway and not via intracellular receptors like MDA-5 and RIG-I.
Reversal of ASFV-ORF-I329L-mediated inhibition of the activation of NFκB and IFN-β promoters by overexpression of TRIF
Activation of the TLR3-TRIF signaling pathway not only induces cytokines, co-stimulatory molecules and antimicrobial peptides that are induced by all TLRs but also antiviral type I interferon and specific chemokines including IP-10 and CCL5 [28, 29].
A direct effect of I329L on TRIF signaling was demonstrated using HEK-TLR3 cells transfected with TRIF as, in the presence of I329L, the IFN-β signaling pathway was inhibited (Fig. 6A). To further investigate the possible impact of I329L on TRIF signaling, HEK-TLR3 cells were simultaneously transfected with I329L (pcDNA3-I329L), the IFN-β luciferase reporter plasmid (IFN-β) and increasing quantities of TRIF plasmid (TRIF) and then stimulated with poly (I:C). As can be seen, overexpression of TRIF reversed the inhibition of reporter activation induced by I329L in a dose-dependent manner, consistent with the hypothesis that the ASFV gene I329L targets TRIF signaling (Fig. 6B).
I329L inhibits activation of the IFN-β promoter at the level of TRIF. 6A) I329L inhibits TRIF-mediated activation of IFN-β. Activation of the IFN-β promoter reporter plasmid was induced via ectopic expression of TRIF in HEK-293T cells through transfection with the TRIF plasmid vector. Transfections were performed with equal amounts of DNA comprising 25 ng of TRIF plasmid vector (TRIF) in the presence of increasing amounts of I329L plasmid vector (100–300 ng) (pcDNA3-I329L) jointly with IFN-β-reporter plasmids (IFN-β). 6B) Overexpression of TRIF reverses I329L-mediated inhibition of IFN-β activation. TRIF was ectopically expressed in HEK-293T cells with increasing amounts of TRIF plasmid vector (25–100 ng) (TRIF) in the presence of 200 ng of the plasmid vector coding for I329L (pcDNA3-I329L) simultaneously with 100 ng of IFN-β-reporter plasmid vector (IFN-β) plus 25 ng of β-gal plasmid vector (β-gal) and after stimulating for 6 hours with 25 μg/ml of poly (I:C) where indicated, before harvesting the cells. In all cases, luciferase activity was measured after 48 h. Luciferase activity was normalized to the β-galactosidase activity obtained with the cotransfected β-gal plasmid internal control. Standard deviations are shown by error bars. Details are in Materials and methods
Discussion
The necessity to recognize and destroy invading pathogens has played a crucial role in the evolution of the immune system of both vertebrates and invertebrates. At the same time, pathogens and, in particular, viruses have evolved reciprocal strategies to manipulate the immune system.
Here, we describe a novel gene from African swine fever virus (ASFV), I329L, which is able to modulate one of the most critical steps of the innate immune response by interfering with TLR3-stimulated activation. This may constitute an advantage for the virus with an impact on both its vertebrate and invertebrate hosts, pigs and ticks, respectively. Conventional bioinformatic analysis of the sequence of I329L predicted a putative type 1 transmembrane protein with an intracellular domain followed by a transmembrane region and an extracellular domain with nine potential glycosylation sites and leucine-rich repeats (LRRs). Both TLR3 and I329L are type I transmembrane proteins with 20% of their sequence intracellular and more than 70% in a highly glycosylated extracellular domain with LRRs. Interestingly, more detailed analysis revealed that the intracellular domain of I329L contains a region of homology with BOX1 and BOX2 of TLR3-TIR domain (Fig. 1A–C). In addition, the putative intracellular domain contains a number of intracellular signaling sequences such as SH2 and SH3 domain binding motifs, a major TRAF2-binding consensus motif and a CK2 phosphorylation site (data not shown). In the work mentioned above, we confirmed that ORF I329L codes for a highly glycosylated protein (Fig. 2C). Significantly, TLR3 is one of the most heavily glycosylated of the TLRs [31], and ORF I329L was demonstrated to be expressed in the cell membranes (Fig. 2A, B) and at the cell surface (Fig. 2D), locations where many receptor and adaptor molecules involved in innate immunity have been identified [32]. Consistent with this, we showed that I329L inhibited double-stranded-RNA-stimulated activation of NFκB (Fig. 4A) and IRF3 (Fig. 4B), two key players in the innate antiviral response. As might be predicted, expression of I329L protein also inhibited the activation of CCL5 (Fig. 5A) and IFN-β (Fig. 5B). Finally, IRF3 activation mediated by expression of TRIF was inhibited by I329L (Fig. 6A), whereas overexpression of TRIF reverted the inhibition of reporter activation induced by I329L in a dose-dependent manner (Fig. 6B). Thus, I329L may be targeting at the level of TRIF, a key adaptor molecule in the MyD88-independent pathway. Direct biochemical proof of this has not been possible, despite many attempts.
Although this is the first ASFV gene manipulating TLR responses to be described, vaccinia virus has evolved two proteins, A46R and A52R, that are inhibitors of the TIR-mediated immune response [12].
Based on our experimental data and the bioinformatic data presented above, some hypothetical models for the ASFV TLR-based strategies to evade the host defense by targeting TLR3 were considered. The most immediate explanation focuses on the intracellular domain of I329L, with its abundance of signaling motifs—in particular, the putative TIR homologous region—and the reversion of I329L-mediated inhibition by overexpression of TRIF. The latter experiment suggested that the cytoplasmic tail of I329L might interfere with recruitment of TRIF, an important adaptor protein involved in the MyD88-independent pathway. Similarly, a direct interaction between the cytoplasmic tail of I329L and TLR3 was suggested by the observed sequence homology between BOX1 of the TIR domain and the intracellular region YIPLIPSY sequence of I329L (Fig. 1B). This conclusion is supported by the experimental data showing I329L-mediated inhibition of IRF3 and NFκB activation and the predicted inhibition of induction of expression of CCL5 (Fig. 5A) and IFN-β (Fig. 5B) induced by externally, but not by internally, delivered poly (I:C) (Fig. 5C). Finally, the observation that increasing amounts of I329L were able to inhibit TRIF-mediated NFκB (Fig. 4A) and IRF3 (Fig. 4B) activation is consistent with the hypothesis that I329L acts at the level of TRIF in the signal transduction pathway. Thus, the possibility that I329L protein interacts with the specific intracellular area responsible for TLR3 homodimerization cannot be excluded, although in most but not all cells, TLR3 is localized to the endosome and not on the cell surface. It is worth adding that when the intracellular domain of ORF I329L is aligned to several TIR domains, it shows a higher homology to the TLR3-TIR domain than to other TIR-domain-containing molecules.
The absence of I329L RNA in ASFV-infected cells until 20 hours postinfection, as shown by northern blot analysis [23], is difficult to reconcile with our findings; that is, a virus mechanism such as I329L, evolved to inhibit IFN and chemokine induction, might be predicted to be expressed early in virus infection. Thus, it will be important to measure expression of I329L by a method that is more sensitive than northern blotting.
In conclusion, the results presented show that ORF I329L is able to impair the cellular responses controlled by TLR3 that lead to both IFN-β production and NFκB activation. The precise mechanism for this inhibition remains to be elucidated, but our current working hypothesis based on our observations and the current literature is that the ORF-I329L-mediated inhibition is intracellular and might be related to an interaction with TRIF. In view of the major impact of I329L on innate immunity, its deletion may provide a rational strategy for construction of an attenuated deletion mutant vaccine.
Abbreviations
- AP-1:
-
activator protein-1
- ASFV:
-
African swine fever virus
- ATF2:
-
activating transcription factor 2
- CCL5:
-
chemokine C-C motif ligand 5
- C/EBP:
-
CCAAT/enhancer-binding proteins
- CRE:
-
cAMP-responsive element
- dsRNA:
-
double-stranded RNA
- ER:
-
endoplasmic reticulum
- GAS:
-
gamma interferon activation site
- HCV:
-
hepatitis C virus
- HRP:
-
horseradish peroxidase
- IFN:
-
interferon
- IFN-β:
-
interferon-β
- IKK:
-
IκB kinases
- IPS-1:
-
interferon promoter stimulator-1
- IRF3:
-
interferon regulatory factor 3
- IRF5:
-
interferon regulatory factor 5
- IRF7:
-
interferon regulatory factor 7
- LRR:
-
leucine-rich repeats
- MDA-5:
-
melanoma differentiation-associated gene 5
- MyD88:
-
myeloid differentiation primary response gene (88)
- NFAT:
-
nuclear factor of activated T-cells
- NFκB:
-
nuclear factor κB
- poly (I:C):
-
polyinosinic:polycytidylic acid
- PRD2:
-
positive regulatory domain 2
- PRR:
-
pathogen recognition receptor
- RIG-I:
-
retinoic-acid-inducible protein I
- SARM:
-
Sterile-alpha and Armadillo motif-containing protein
- STAT:
-
signal transducer and activator of transcription
- TIR:
-
Toll/IL1 receptor
- TLR:
-
Toll-like receptor
- TNF-α:
-
tumor necrosis factor-α
- TRAF2:
-
TNF receptor-associated factor 2
- TRIF:
-
TIR-domain-containing adaptor-inducing interferon-β
References
Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D et al (2004) African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol 100:117–134
Chapman DA, Tcherepanov V, Upton C, Dixon LK (2008) Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J Gen Virol 89:397–408
Kawai T, Akira S (2007) Antiviral signaling through pattern recognition receptors. J Biochem 141:137–145
Brikos C, O’Neill LA (2008) Signalling of toll-like receptors. Handb Exp Pharmacol 183:21–50
Watters TM, Kenny EF, O’Neill LA (2007) Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol 85:411–419
Jin MS, Lee JO (2008) Structures of TLR–ligand complexes. Curr Opin Immunol 20:414–419
O’Neill LA, Bowie AG (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 7:353–364
Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K et al (2002) Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol 169:6668–6672
Kaisho T, Akira S (2006) Toll-like receptor function and signaling. J Allergy Clin Immunol 117:979–987
Hiscott J (2007) Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev 18:483–490
Alcami A, Koszinowski UH (2000) Viral mechanisms of immune evasion. Trends Microbiol 8:410–418
Bowie AG, Unterholzner L (2008) Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922
Vercammen E, Staal J, Beyaert R (2008) Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev 21:13–25
Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O’Neill LA (2000) A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc Natl Acad Sci USA 97:10162–10167
Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW et al (2003) The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med 197:343–351
Maloney G, Schroder M, Bowie AG (2005) Vaccinia virus protein A52R activates p38 mitogen-activated protein kinase and potentiates lipopolysaccharide-induced interleukin-10. J Biol Chem 280:30838–30844
Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC et al (2005) Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med 201:1007–1018
Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M et al (2005) Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA 102:2992–2997
Li XD, Sun L, Seth RB, Pineda G, Chen ZJ (2005) Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 102:17717–17722
Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG (2006) The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol 7:1074–1081
Yang Y, Huang CT, Huang X, Pardoll DM (2004) Persistent Toll-like receptor signals are required for reversal of regulatory T cell-mediated CD8 tolerance. Nat Immunol 5:508–515
Zhu J, Martinez J, Huang X, Yang Y (2007) Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood 109:619–625
Rodriguez JM, Salas ML, Vinuela E (1992) Genes homologous to ubiquitin-conjugating proteins and eukaryotic transcription factor SII in African swine fever virus. Virology 186:40–52
Sonnhammer EL, von HG, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182
nett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70:611–625
Marshall-Clarke S, Downes JE, Haga IR, Bowie AG, Borrow P, Pennock JL et al (2007) Polyinosinic acid is a ligand for toll-like receptor 3. J Biol Chem 282:24759–24766
Medzhitov R, Preston-Hurlburt P, Janeway CA Jr (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394–397
Takeuchi O, Hemmi H, Akira S (2004) Interferon response induced by Toll-like receptor signaling. J Endotoxin Res 10:252–256
Melchjorsen J, Sorensen LN, Paludan SR (2003) Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J Leukoc Biol 74:331–343
Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E et al (2003) LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med 198:1043–1055
Bell JK, Askins J, Hall PR, Davies DR, Segal DM (2006) The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci USA 103:8792–8797
Fan S, Chen S, Liu Y, Lin Y, Liu H, Guo L et al (2008) Zebrafish TRIF, a Golgi-localized protein, participates in IFN induction and NF-kappaB activation. J Immunol 180:5373–5383
Acknowledgments
This work was supported with research grants from the Fundação para a Ciência e Tecnologia (FCT) (POCTI/MGI/45100/2002), the Wellcome Trust (WT075813MA) and the EU (QLK3-CT-2000-00362). V.L.O was a student from the Gulbenkian Ph.D. Program in Biomedicine and a recipient of a FCT fellowship (SFRH/BD/9617/2002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that they have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Additional information
V. L. de Oliveira and S. C. P. Almeida contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Oliveira, V.L., Almeida, S.C.P., Soares, H.R. et al. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch Virol 156, 597–609 (2011). https://doi.org/10.1007/s00705-010-0894-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0894-7