Abstract
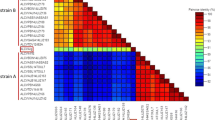
The complete genome sequence for an isolate of the Ugandan and Tanzanian strain types of Cassava brown streak virus have been determined using the novel approach of non-directed next generation sequencing. Comparison of the genome sequences revealed that CBSV is highly heterogeneous at the isolate level as well as the strain level. The isolate of the Ugandan strain was found to have a genome 9,070 nucleotides long coding for a polypeptide with 2,902 amino acid residues. The isolate of the Tanzanian strain was 9,008 nucleotides long and coded for a polypeptide with 2,916 amino acid residues. Nucleotide identity between the isolates across the genome was 76%, with protein encoding regions 57–77% and individual proteins had 65–91% amino acid similarity. In addition between the two strains four protein products (PIPO, CI, NIa-Vpg and coat protein) varied in size and an unusual HAM1-like protein, whilst of identical nucleotide length, was found to have the lowest homology. The implication of diversity of CBSV is discussed in the context of speciation, evolution, development of diagnostics, and breeding for resistance.

Similar content being viewed by others
References
Adams IP, Glover RH, Monger WA, Mumford R, Jackeviciene E, Navalinskiene M, Samuitien M, Boonham N (2009) Next generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol Plant Pathol 10:537–545
Adams MJ, Antoniw JF, Fauquet CM (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol 150:459–479
Alicai T, Omongo CA, Maruthi MN, Hillocks RJ, Baguma Y, Kawuki R, Bua A, Otim-Nape GW, Colvin J (2007) Re-emergence of cassava brown streak disease in Uganda. Plant Dis 91(1):24–29
Chung BYW, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Nat Acad Sci 105(15):5897–5902
Hillocks RJ, Jennings DK (2003) Cassava brown streak disease: a review of present knowledge and research needs. Int J Pest Manag 49:225–234
Mbanzibwa DR, Tian Y, Mukasa SB, Valkonen JPT (2009) Cassava brown streak virus (Potyviridae encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC-Pro. J Virol 83(13):6934–6940
Mbanzibwa DR, Tian Y, Tugume AK, Mukasa SB, Tairo F, Kyamanywa S, Kullaya A, Valkonen JPT (2009) Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch Virol 154:353–359
Monger WA, Seal S, Issac AM, Foster GD (2001) Molecular characterization of Cassava brown streak virus coat protein. Plant Pathol 50:527–534
Monger WA, Seal S, Cotton S, Foster GD (2001) Identification of different isolates of Cassava brown streak virus and development of a diagnostic test. Plant Pathol 50:768–775
Shukla DD, Frenkel MJ, Ward CW (1991) Structure and function of the potyvirus genome with special reference to the coat protein coding region. Can J Plant Pathol 13:178–191
Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) (2005) Virus taxonomy, eighth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam, p 822, p 830
Acknowledgments
The authors wish to acknowledge financial support from the Bill and Melinda Gates Foundation and thank the numerous individuals involved in the Great Lakes Cassava Initiative, from which some of the infected cassava material reported in this paper was obtained. We also wish to thank Dr M. N. Maruthi (NRI, Greenwich University, UK) for providing additional positive material against which findings could be checked.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Monger, W.A., Alicai, T., Ndunguru, J. et al. The complete genome sequence of the Tanzanian strain of Cassava brown streak virus and comparison with the Ugandan strain sequence. Arch Virol 155, 429–433 (2010). https://doi.org/10.1007/s00705-009-0581-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0581-8