Abstract
The family Totiviridae comprises viruses with nonsegmented dsRNA genomes and isometric virions. A new genus, Victorivirus, has been approved for this family, named from the specific epithet of Helminthosporium victoriae, host of the type species, Helminthosporium victoriae virus 190S. Distinguishing characteristics of the 11 viruses so far assigned to this genus include infection of filamentous fungi, an apparently coupled termination–reinitiation mechanism for translating the RNA-dependent RNA polymerase as a separate product from the upstream capsid protein, and sequence-based phylogenetic grouping in a distinct clade from other family members.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Totiviridae encompasses a broad range of viruses characterized by isometric virions, ~40 nm in diameter, that each contain a nonsegmented dsRNA genome coding in most cases for only a capsid protein (CP) and an RNA-dependent RNA polymerase (RdRp) [9]. Viruses of the 18 species assigned to the genera in this family as of spring 2008 by the International Committee on Taxonomy of Viruses (ICTV) persistently infect either fungi or protozoa, although a similar virus causing disease in penaeid shrimp has been reported [25]. To date, only three genera have been formally recognized: Totivirus, Giardiavirus, and Leishmaniavirus. Viruses infecting yeast, smut, or filamentous fungi have been placed in the genus Totivirus, and ones infecting parasitic protozoa in the latter two genera.
At least three different strategies for RdRp expression appear to be used among members of this family: (1) as a fusion with CP (CP/RdRp) consequent to ribosomal frameshifting, as in Saccharomyces cerevisiae viruses L-A (ScV-L-A) and L-BC and also among the viruses that infect parasitic protozoa [5, 6, 16, 18, 19]; (2) as a fusion with CP without the use of ribosomal frameshifting, as in Ustilago maydis virus H1, in which case the RdRp is putatively released from the fusion by proteolysis [17]; and—of prime relevance to this article—(3) as a separate protein consequent to an apparently coupled termination–reinitiation mechanism, as in Helminthosporium victoriae virus 190S (HvV190S) [14, 29] and proposed for other viruses that infect filamentous fungi [2, 9, 21, 23, 24, 26, 27, 32, 35]. Sequence-based analyses have shown that the HvV190S-like viruses moreover define a discrete phylogenetic clade [1, 2, 9, 21, 27].
The distinguishing characteristics of the HvV190S-like viruses led us to propose a new genus to encompass them. Establishment of this genus, named Victorivirus from the specific epithet of H. victoriae, host of the type species, Helminthosporium victoriae virus 190S, was considered by the Executive Committee of the ICTV at its 2007 midterm meeting in Kingston, ON, Canada. The revised proposal was then approved by that committee at its 2008 meeting in conjunction with the ICTV plenary session in Istanbul, Turkey. It now awaits ratification by the full ICTV membership.
Taxonomic structure
Order: | Unassigned |
Family: | Totiviridae |
Genus: | Victorivirus |
Type species: | Helminthosporium victoriae virus 190S |
Other species: | Chalara elegans RNA virus 1 |
Coniothyrium minitans RNA virus | |
Epichloe festucae virus 1 | |
Gremmeniella abietina RNA virus L1 | |
Helicobasidium mompa totivirus 1–17 | |
Magnaporthe oryzae virus 1 | |
Sphaeropsis sapinea RNA virus 1 | |
Sphaeropsis sapinea RNA virus 2 | |
Tentative species: | Botryotinia fuckeliana totivirus 1 |
Magnaporthe oryzae virus 2 |
Helminthosporium victoriae virus 190S was previously classified in the genus Totivirus and was the only ICTV-recognized species among those listed above. In association with approving the genus Victorivirus, the ICTV Executive Committee reassigned it as type species of this new genus and also approved the other listed species. Although H. victoriae virus 190S is its ICTV-approved name used in this article and abbreviated HvV190S, it is commonly referred to in the literature as H. victoriae 190S virus and abbreviated Hv190SV. Two viruses, B. fuckeliana totivirus 1 and Magnaporthe oryzae virus 2, were not included in the initial proposal to the ICTV because pertinent sequencing reports [4, 21] were not yet available. They are listed above as tentative species and will be included in a future ICTV proposal to create new species in an existing genus. Gremmeniella abietina RNA virus L2 [33] is another reported strain of species Gremmeniella abietina RNA virus L1 [32].
Biological properties
There are no known, dedicated vectors in nature for the transmission of members of the genus Victorivirus (victoriviruses). Neither is extracellular transmission in the absence of a vector known to occur in any regular manner. Instead, these viruses are transmitted intracellularly: vertically during host cell division and sporogenesis and horizontally during cell–cell fusion such as in hyphal anastomosis between compatible host strains [9]. Victoriviruses that infect ascomycetous fungi appear to be largely excluded during ascospore formation.
Experimental host ranges for victoriviruses are unknown because of a lack of suitable infectivity assays. Their natural host ranges are limited to individuals within the same or closely related vegetative compatibility groups. They are disseminated in nature via asexual spores, which are often produced in great profusion.
Victoriviruses are associated with persistent, commonly symptomless infections of their hosts. Although several of these host fungi are phytopathogens, there are so far no confirmed examples in which fungal pathogenicity is positively or negatively modulated by victorivirus infection. Whether HvV190S affects the pathogenicity of H. victoriae, the causative agent of Victoria blight of oats, remains under investigation [9]. Unlike in yeast and smut totiviruses [34], there are no known examples of victoriviruses that express killer toxins from satellite dsRNAs.
Mixed infections with two or more fungal viruses are common, probably as a consequence of how they are transmitted in nature and replicate persistently in their hosts. Dual infection of Sphaeropsis sapinea with Sphaeropsis sapinea RNA viruses 1 and 2 is an example of a mixed infection involving two distinct victoriviruses [26]. Dual infection of H. victoriae with victorivirus HvV190S and chrysovirus H. victoriae 145S virus is an example of a mixed infection involving two dsRNA viruses from different families [13, 28].
The cell biology of victorivirus infections, including subcellular localizations of their replication and assembly complexes, remains largely unexplored. Except for the multifunctional cellular protein Hv-p68, a protein kinase/alcohol oxidase/RNA-binding protein that is overproduced in HvV190S-infected H. victoriae isolates [30, 36], host factors that are required for, modulate, inhibit, or are affected by victorivirus infections are also unknown.
Virions and replication
Victoriviruses have isometric virions, 35–45 nm in diameter as visualized by negative staining and transmission electron microscopy. The capsids appear single layered and thin, with relatively smooth surfaces. Genome-deduced protein sequences have suggested that the CPs of different victoriviruses are similarly sized (Table 1), from 746 to 789 aa or approximately 77 to 83 kDa. The victorivirus CPs are unique in the family Totiviridae in having an Ala/Gly/Pro-rich region predicted near their C termini. The C-terminal sequence of HvV190S CP, for example, is VPLPPAPGAAPPPPPGPPNGPPAGPPPSDDGSSNPAAPVPTAIHAPPAAAQADRAEGQ [14]. HvV190S is the best characterized of these viruses with regard to its molecular features.
The capsid structure of HvV190S has been determined at a nominal resolution of 14 Å by using transmission electron cryomicroscopy and three-dimensional image reconstruction [1]. The capsid is made up of 60 asymmetric CP dimers arranged in a T = 1 (so-called “T = 2”) icosahedral lattice. This capsid organization is very much like that of ScV-L-A [3, 22] and is indeed related to that shared by members of several other dsRNA virus families. The outer surface of the capsid is notably smooth except for short projections that appear to coalesce over the fivefold axes. The buoyant density in CsCl of HvV190S virions is 1.43 g/cm3, and (as its name implies) the sedimentation coefficient is 190S (S20, w, in Svedberg units). Particles lacking RNA have a sedimentation coefficient of ~80S [1, 28].
Full-length HvV190S CP migrates with a relative molecular weight of 88,000 in denaturing gels. Two C-terminally cleaved forms of CP, detected as 83,000 and 78,000 gel bands, can also be associated with HvV190S particles [10, 15], but are not required for capsid assembly [31]. The C-terminal 132 aa of CP are in fact dispensable for capsid assembly [15]. The two larger forms of CP are phosphorylated, probably near their C termini since the smallest form is not [12]. Phosphorylated and/or cleaved forms of CP may be important for the RNA packaging and synthesis functions of HvV190S particles [9, 12].
Genome-deduced protein sequences have suggested that the RdRps of different victoriviruses are also similarly sized (Table 1), from 825 to 871 aa or approximately 90 to 98 kDa. A few, perhaps one or two, copies of the RdRp have been shown to be packaged in HvV190S virions [14], probably by noncovalent anchoring inside the capsid [1]. These virion-associated RdRp molecules are expected to mediate synthesis of the victorivirus RNAs during replication and transcription.
The genome of victoriviruses is a nonsegmented molecule of dsRNA, consistently sized near 5,000 bp in nondenaturing gels. As demonstrated for HvV190S [14], the genomic plus strand of each victorivirus is thought to be neither capped at its 5′ end nor polyadenylylated at its 3′ end. A single one of these dsRNA molecules is thought to be packaged in the capsid-enclosed interior of each virion, along with the few copies of RdRp. In many fungal viruses, satellite or defective RNAs can be packaged and replicated in parallel with the viral genome segment(s). Their significance to infection is commonly unknown, but in yeast and smut totiviruses, satellite dsRNAs can encode killer toxins that provide an advantage to the infected host [34]. To date, satellite or defective dsRNAs have not been widely reported in victoriviruses, although a putative satellite dsRNA has been described in association with Helicobasidium mompa totivirus 1–17 [23].
Like those of other dsRNA viruses, the RdRp-containing particles of victoriviruses are expected to be nanomachines for RNA synthesis, as already shown for HvV190S. Transcription by HvV190S virions is asymmetric (meaning that only plus-strand transcripts are generated) and conservative (meaning that the parental plus strand is retained as part of the duplex template while the newly synthesized plus strand is released) [11]. Moreover, as demonstrated for HvV190S [11], the primary products of transcription by each victorivirus are thought to be full-length copies of the genomic plus strand, which serve for translation by host ribosomes and also for incorporation into newly assembling virions. Since the genomic plus strand of HvV190S is not capped or polyadenylylated, these transcripts of HvV190S and other victoriviruses are presumably not as well. Whether the capsids of HvV190S and other victoriviruses function to cleave the 5′ caps from host mRNAs, as does ScV-L-A [22], remains unknown. The victorivirus RdRp is also presumed to mediate one round of minus-strand synthesis (replication) to generate the duplex genome molecule in newly assembled virions before switching to the transcription mode. Since victoriviruses are regularly transmitted only by intracellular routes, their virions are expected to lack the machinery for cell entry.
Genomic and coding properties
Complete nucleotide sequences of HvV190S and the ten other viruses so far assigned to the genus Victorivirus have been deposited in GenBank (Table 1). All have total lengths of 4,975–5,359 bp. There is no clear evidence for widely conserved sequences at the genome termini of the different victoriviruses.
The genomic plus strand of each victorivirus has two long open reading frames (ORFs). The upstream ORF (ORF1) encodes the CP, the downstream ORF (ORF2) encodes the RdRp, and the two ORFs are in different frames (Fig. 1). In 8 of the 11 victoriviruses including HvV190S, ORF2 is in the −1 frame relative to ORF1, and in all but one of the 11 viruses (Magnaporthe oryzae virus 1 [35] is the exception), the ORFs overlap by just a few nt (see further description below). Among the 11 victoriviruses, the 5′ untranslated region (UTR) varies fairly widely in length, from 61 to 574 nt, although in nine of the viruses including HvV190S, the range is less, from 199 to 336 nt (Table 1). The 3′ UTR is more consistently sized, from 43 to 114 nt, among the 11 victoriviruses. The genomic minus strand of each virus appears to have little or no coding potential, consistent with its lack of access to host ribosomes during the viral replication cycle.
The sequence region in which the end of ORF1 overlaps or approaches the beginning of ORF2 has related features in all victoriviruses. In 7 of the 11 viruses including HvV190S, the AUG start codon at the beginning of ORF2 overlaps the stop codon at the end of ORF1 in the tetranucleotide sequence AUGA. In three other victoriviruses, the AUG start codon at the beginning of ORF2 precedes the stop codon at the end of ORF1 by two intervening nt (e.g., AUGUCUAG in Chalara elegans RNA virus 1 [24]). In the one remaining virus, M. oryzae virus 1, the AUG start codon at the beginning of ORF2 follows the stop codon at the end of ORF1 by two intervening nt (UAGAUAUG). Thus, despite some variation, the predicted ORF1 stop and ORF2 start codons are closely juxtaposed in each genome, which is thought to contribute to the internal initiation mechanism for ORF2 translation from the dicistronic transcript.
The genomic and coding properties of HvV190S are the best characterized among victoriviruses. Several lines of evidence support the conclusion that expression of HvV190S CP and HvV190S RdRp initiate from the AUG start codons at positions 290–292 and 2605–2607, respectively [14, 15, 29]. In addition, the UGA stop codon for ORF1 at nt 2,606–2,608 has been verified by site-directed mutagenesis. As also described above, the RdRp-encoding ORF2 of HvV190S is downstream of ORF1, and its start codon (nt 2,605–2,607) overlaps the stop codon of ORF1 (nt 2,606–2,608) in the tetranucleotide sequence AUGA (Fig. 1). HvV190S RdRp is detectable as a separate, virion-associated component, and not as part of a larger fusion protein, consistent with its independent translation from ORF2 [14]. Its translation as an independent protein from dicistronic transcripts in eukaryotic cells has also been demonstrated and appears to depend on one or more host factors since its translation from matching transcripts in bacterial cells is negligible [29]. Nevertheless, its efficiency of translation in eukaryotic cells is substantially less than that of the CP, consistent with the many fewer copies of RdRp in HvV190S virions [14]. The 5′ UTR of the HvV190S plus strand is predicted to be highly structured and contains two minicistrons [14]. Combined with the fact that the genomic plus strand is not capped or polyadenylylated, these features suggest that the CP-encoding ORF1 (with its start codon present in suboptimal context according to Kozak criteria) is translated via a cap-independent mechanism (Fig. 1).
Phylogenetic relationships
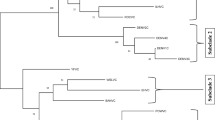
HvV190S and other victoriviruses are phylogenetically closer to each other than to the family members from yeast and smut fungi, which remain grouped in the genus Totivirus. The fact that independent alignments of the CP and RdRp sequences suggest similar relationships among these groups (Fig. 2) supports the conclusion that the HvV190S-like viruses should reside in a separate genus. In fact, nearly all other members of the family Totiviridae, including the yeast and smut totiviruses, express the RdRp as part of a polyprotein [8], and this key coding difference parallels their separation from victoriviruses in the sequence-based phylogenies. The different patterns of phylogenetic branching in the two trees in Fig. 2 are probably explained in part by different rates of evolution between the CP and the RdRp in these viruses.
Neighbor-joining phylogenetic trees (bootstrap consensus) based on the complete CP sequences (left) or a 480-aa sequence region encompassing the RdRp conserved motifs (right) of each analyzed virus. RdRp conserved motifs 1–8 were as previously designated [6]. Sequences were multiply aligned using PRANK+F (Prankster version 080215) [20]. Trees were generated using PAUP* (version 4.0b10) and plotted using TreeEdit (version 1.0a10). Bootstrap values (%) calculated from 2,000 replicates are indicated at the nodes. See Table 1 for abbreviations and GenBank (GB) accession numbers for viruses in genus Victorivirus. Other recognized or tentative members of family Totiviridae included in the tree are: EbRV1, Eimeria brunetti RNA virus 1 (GB, AF356189); LRV1-1, Leishmania RNA virus 1-1 (GB, M92355); LRV2-1, Leishmania RNA virus 2-1 (GB, U32108), LRV1-4, Leishmania RNA virus 1-4 (GB, U01899); TVV1, Trichomonas vaginalis virus 1 (GB, U08999); TVV2, Trichomonas vaginalis virus II (GB, AF127178); TVV3, Trichomonas vaginalis virus 3 (GB, AF325840); ScV-L-A, Saccharomyces cerevisiae virus L-A (GB, J04692); ScV-L-BC, Saccharomyces cerevisiae virus L-BC (GB, U01060); UmV-H1, Ustilago maydis virus H1 (GB, U01059); GLV, Giardia lamblia virus (GB, L13218); and IMNV, penaeid shrimp infectious myonecrosis virus (GB, AY570982). The trees were rooted by designating IMNV as outgroup. Viruses in genus Victorivirus are labeled in orange (including tentative members BfTV1 and MoV2), those in genus Leishmaniavirus in green, those in genus Giardiavirus in magenta (including tentative members TVV1, TVV2, and TVV3), and those in genus Totivirus in cyan. Viruses labeled in black (EbRV1 and IMNV) are tentative members of family Totiviridae, not yet assigned to a genus
Interestingly, according to phylogenetic analyses of both CP and RdRp sequences, victoriviruses are more closely related to leishmaniaviruses than to the viruses of yeast and smut fungi (Fig. 2). For example, whereas HvV190S RdRp shares 34% sequence identity with Leishmania RNA virus 1-1 across a 480-aa region encompassing the eight conserved RdRp motifs [8], its identity with ScV-L-A in that region is only 22%. The closer relationship of victoriviruses to leishmaniaviruses was previously noted [8] and supports the conclusion that fungal viruses in the family Totiviridae are polyphyletic.
Another reported virus from a parasitic protozoan, Eimeria brunetti RNA virus 1 (EbRV1) [7], is identified in the phylogenetic trees as the virus next most closely related to victoriviruses (Fig. 2). Notably, EbRV1 appears to share the victorivirus strategy for RdRp expression as a separate protein, with the RdRp start codon overlapping the CP stop codon in the −1 frame at an AUGA tetranucleotide. In addition, the lengths of the EbRV1 genome (5,358 nt), 5′ UTR (357 nt), and CP (770 aa) are all within the ranges for these values defined by assigned victoriviruses (see Table 1). Because of the distinct host from which EbRV1 was isolated, however, we have not grouped it in the genus Victorivirus. Instead, depending on future results, EbRV1 might be more properly placed in another genus of protozoan viruses.
The phylogenetic trees constructed with CP and RdRp sequences in this and other reports [1, 9, 21, 27] suggest the existence of two subclades of viruses within the genus Victorivirus (Fig. 2). Because other known properties do not differentiate these subclades, however, all have been encompassed by the new genus.
References
Castón RJ, Luque D, Trus BL, Rivas G, Alfonso C, González JM, Carrascosa JL, Annamalai P, Ghabrial SA (2006) Three-dimensional structure and stoichiometry of Helminthosporium victoriae 190S totivirus. Virology 347:323–332
Cheng J, Jiang D, Fu Y, Li G, Peng Y, Ghabrial SA (2003) Molecular characterization of a dsRNA totivirus infecting the sclerotial parasite Coniothyrium minitans. Virus Res 93:41–50
Cheng RH, Caston JR, Wang GJ, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, Steven AC (1994) Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol 244:255–258
De Guido MA, Minafra A, Santomauro A, Pollastro S, De Miccolis RM, Faretra F (2007) Molecular characterization of mycoviruses from Botryotinia fuckeliana. GenBank, Accession No. NC_009224
Diamond ME, Dowhanick JJ, Nemeroff ME, Pietras DF, Tu C-L, Bruenn JA (1989) Overlapping genes in a yeast dsRNA virus. J Virol 63:3983–3990
Dinman JD, Icho T, Wickner RB (1991) A-1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag–pol fusion protein. Proc Natl Acad Sci USA 88:174–178
Fraga JS, Katsuyama AM, Fernandez S, Madeira AMBN, Briones MRS, Gruber A (2006) The genome of the Eimeria brunetti RNA virus 1 is more closely related to fungal than to protozoan viruses. GenBank, Accession No. NC_002701
Ghabrial SA (1998) Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16:119–131
Ghabrial SA (2008) Totiviruses. In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of virology vol 5, 3rd edn. Elsevier, Oxford, pp 163–174
Ghabrial SA, Bibb JA, Price KH, Havens WM, Lesnaw JA (1987) The capsid polypeptides of the 190S virus of Helminthosporium victoriae. J Gen Virol 68:1791–1800
Ghabrial SA, Havens WM (1989) Conservative transcription of Helminthosporium victoriae 190S virus dsRNA in vitro. J Gen Virol 70:1025–1035
Ghabrial SA, Havens WM (1992) The Helminthosporium victoriae 190S mycovirus has two forms distinguishable by capsid protein composition and phosphorylation state. Virology 188:657–665
Ghabrial SA, Soldevila AI, Havens WM (2002) Molecular genetics of the viruses infecting the plant pathogenic fungus Helminthosporium victoriae. In: Tavantzis S (ed) Molecular biology of double-stranded RNA: concepts and applications in agriculture, forestry and medicine. CRC Press, Boca Raton, pp 213–236
Huang S, Ghabrial SA (1996) Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci USA 93:12541–12546
Huang S, Soldevila AI, Webb BA, Ghabrial SA (1997) Expression, assembly and proteolytic processing of Helminthosporium victoriae 190S totivirus capsid protein in insect cells. Virology 234:130–137
Icho T, Wickner RB (1989) The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem 264:6716–6723
Kang J, Wu J, Bruenn JA, Park C (2001) The H1 double-stranded RNA genome of Ustilago maydis virus-H1 encodes a polyprotein that contains structural motifs for capsid polypeptide, papain-like protease, and RNA-dependent RNA polymerase. Virus Res 76:183–189
Lee SE, Suh JM, Scheffter S, Patterson JL, Chung IK (1996) Identification of a ribosomal frameshift in Leishmania RNA virus 1–4. J Biochem 120:22–25
Li L, Wang AL, Wang CC (2001) Structural analysis of the −1 ribosomal frameshift elements in giardiavirus mRNA. J Virol 75:10612–10622
Löytynoja A, Goldman N (2008) Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320:1632–1635
Maejima K, Himeno M, Komatsu K, Kakizawa S, Yamaji Y, Hamamoto H, Namba S (2008) Complete nucleotide sequence of a new double-stranded RNA virus from the rice blast fungus, Magnaporthe oryzae. Arch Virol 153:389–391
Naitow H, Tang J, Canady M, Wickner RB, Johnson JE (2002) L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol 9:725–728
Nomura K, Osaki H, Iwanami T, Matsumoto N, Ohtsu Y (2003) Cloning and characterization of a totivirus double-stranded RNA from the plant pathogenic fungus, Helicobasidium mompa Tanaka. Virus Genes 23:219–226
Park Y, James D, Punja ZK (2005) Co-infection by two distinct totivirus-like double stranded RNA elements in Chalara elegans (Thielaviopsis basicola). Virus Res 109:71–85
Poulos BT, Tang KF, Pantoja CR, Bonami JR, Lightner DV (2006) Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J Gen Virol 87:987–996
Preisig O, Wingfield BD, Wingfield MJ (1998) Coinfection of a fungal pathogen by two distinct double-stranded RNA viruses. Virology 252:399–406
Romo M, Leuchtmann A, Garcia B, Zabalgogeazcoa I (2007) A totivirus infecting the mutualistic fungal endophyte Epichloë festucae. Virus Res 124:38–43
Sanderlin RS, Ghabrial SA (1978) Physiochemical properties of two distinct types of virus-like particles from Helminthosporium victoriae. Virology 87:142–151
Soldevila A, Ghabrial SA (2000) Expression of the totivirus Helminthosporium victoriae 190S virus RNA-dependent RNA polymerase from its downstream open reading frame in dicistronic constructs. J Virol 74:997–1003
Soldevila A, Ghabrial SA (2001) A novel alcohol oxidase/RNA-binding protein with affinity for mycovirus double-stranded RNAs from the filamentous fungus Helminthosporium (Cochliobolus) victoriae—molecular and functional characterization. J Biol Chem 276:4652–4661
Soldevila AI, Huang S, Ghabrial SA (1998) Assembly of the Hv190S totivirus capsid is independent of posttranslational modification of the capsid protein. Virology 251:327–333
Tuomivirta TT, Hantula J (2003) Two unrelated double-stranded RNA molecule patterns in Gremmeniella abietina type A code for putative viruses of the families Totiviridae and Partitiviridae. Arch Virol 148:2293–2305
Tuomivirta TT, Hantula J (2005) Three unrelated viruses occur in a single isolate of Gremmeniella abietina var. abietina type A. Virus Res 110:31–39
Wickner RB (2006) Viruses and prions of yeast, fungi, and unicellular organisms. In: Knipe DM, Howley PM (eds) Fields virology, 5th edn. Lippincott Williams & Wilkins, Philadelphia, pp 737–768
Yokoi T, Yamashita S, Hibi T (2007) The nucleotide sequence and genome organization of Magnaporthe oryzae virus 1. Arch Virol 152:2265–2269
Zhao T, Havens WM, Ghabrial SA (2006) The disease phenotype of virus-infected Helminthosporium victoriae is independent of overexpression of the cellular alcohol oxidase/RNA-binding protein Hv-p68. Phytopathology 96:326–332
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghabrial, S.A., Nibert, M.L. Victorivirus, a new genus of fungal viruses in the family Totiviridae . Arch Virol 154, 373–379 (2009). https://doi.org/10.1007/s00705-008-0272-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-008-0272-x