Abstract
Vegetation phenology models still rely on temperature as the primary limiting factor to growth. They generally do not recognize the importance of photoperiod and water availability, which can cause them to under-perform. Moreover, few models have used machine learning algorithms to find relationships in the data. In this paper, four Vegetation Indexes (VIs), namely the green chromatic coordinate (GCC), the vegetation contrast index (VCI), the normalized difference vegetation index (NDVI) and the two-band enhanced vegetation index (EVI2), are predicted for the North American Great Plains. This is possible by using six PhenoCams, Daily Surface Weather and Climatological Summaries (DAYMET), processing them with the machine learning algorithm XGBoost (XGB) and comparing them with seven phenophase stages throughout a growth cycle. Examining the results, GCC was the best fitting model with an R2 of 0.946, while EVI2 was the poorest with an R2 of 0.895. Also, the results indicate that changing temperature and precipitation patterns are driving a significant change in phenology of the grasslands. We developed a model capable of explaining 90 to 93% of the variability in four VIs across six grassland PhenoCam sites over the growing season using the XGB regression. Our model demonstrates the importance of including photoperiod, temperature, and precipitation information when modeling vegetation phenology. Finally, we were able to construct a 38-year phenology record at each PhenoCam location.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Grasslands cover approximately 59 million km2 of the Earth’s surface (Hufkens et al. 2016) making up between 10 and 30% of the global carbon stock (Scurlock and Hall 1998); this makes grasslands the second largest carbon sink after forests (Anderson 1991). In North America, the Great Plains cover approximately 2.9 million km2 within an east-to-west gradient of tall to short-grass prairie. However, the conversion of grassland to cropland has drastically reduced the remaining native prairie ecosystems. In 2018, it was estimated that only half of these grassland ecosystems remain, with 87% of them located on poor and marginal quality soils (World Wildlife Fund 2018). The variation within the Great Plains creates a variety of community types typically dominated by C3 grasses in the north and east (more precipitation and cooler temperatures), and C4 grasses in the south and west (less precipitation and higher temperatures) (Petrie et al. 2016). The C3-pathway for photosynthesis is common in temperate regions in grasses such as wheatgrass (Agropyron), bentgrass (Agrostis), and foxtail (Alopecurus), while the C4-pathway is common in arid regions where the weather is typically hotter and drier with grasses such as bluestem (Bothriochloa), threeawn (Aristida), and grama (Bouteloua) (Jones and Vaughan 2010; Stubbendieck et al. 2017). Along with a large amount of spatial variability, grasslands are also characterized by high amounts of temporal variability (Flanagan and Adkinson 2011). This means that climate change induced shifts in grassland phenology will likely only be detectable using long-term monitoring over several years to decades (Henebry 2013).
Modeled scenarios under forecast future climate conditions suggest that North America will see an increase in both the length of the growing season and the productivity of grasslands, including an earlier onset of spring (Schwartz et al. 2006). This is because the modeled grasslands are expected to become more efficient in retaining moisture under higher CO2 levels, allowing for more efficient use of water and a reduction in the amount of water lost in transpiration (Hufkens et al. 2016). This suggests that precipitation must fall below a threshold before it has a noticeable effect on growing season length (Browning et al. 2017).However, a controlled test of grassland phenology using plants grown within a warmer temperature, elevated CO2, increased nitrogen, and increased precipitation has shown an array of responses that were not all anticipated. For example, additions of CO2 delayed spring greenness while increased nitrogen slowed down plant growth acceleration. Precipitation had no effect, suggesting it was not a limiting factor for the controlled plants, while increased temperature was the only factor to have the expected outcome, causing plants to flower earlier by 2–5 days (Cleland et al. 2006). Field observations of arid grasslands using both PhenoCams (Richardson et al. 2018) as well as satellite imagery are also in agreement that warmer temperatures bring an earlier start of season to the grasslands. But in an arid environment precipitation has been found to influence the recorded vegetation indices (VIs), even causing a second peak of greenness in the growing season after a large precipitation event (Browning et al. 2017).
Identifying the limiting factor for growth of grassland phenology is a challenging task, with factors such as temperature and precipitation fluctuating throughout the growing season to limit plant growth (Wang et al. 2003). Many phenology models still rely on temperature as the primary limiting factor to growth, and because of this they under-perform by not recognizing the importance of photoperiod and water availability (Piao et al. 2019). Temperature-driven models may fail to help predict future phenology patterns from climate change since plants can have a reduced sensitivity to temperature (Fu et al. 2015). Instead, new models should be developed to account for the interactions between the many environmental factors that drive plant growth.
Machine learning has gained traction in Earth sciences and ecology, with many machine learning models outperforming traditional statistical models (Dai et al. 2019). Machine learning algorithms apply non-linear techniques that can often identify complex underlying relationships in the data (Zhang et al. 2019). Regardless of these advantages, there are few phenology models that take advantage of the benefits provided by machine learning (Dai et al. 2019). One recently developed machine learning algorithm, known as XGBoost (XGB), is a gradient boosted decision tree capable of both regression and classification tasks (Chen and Guestrin 2016). Improvements made in XGB make it more robust at handling noise, as well as dealing with unbalanced and skewed datasets (Zhang et al. 2019). This makes it an excellent choice when working with empirical data that often fails to meet the requirements of parametric statistical analysis. However, using machine learning for phenology requires long time series datasets with few data gaps, although, even then, analysis can be challenging when noise is present (Belda et al. 2020).
PhenoCams are digital web-enabled cameras that are capable of imaging ecosystems with high temporal resolution (Richardson 2019). PhenoCams record changes in vegetation throughout the growing season by capturing multiple images per day using the visible and sometime the near-infrared portions of the electromagnetic spectrum. Stages in vegetation phenology are known as phenophases and include greenup in the spring, and senescence in the fall (Richardson and Braswell 2009). Individual images captured by PhenoCams are used to calculate VIs that record changes in vegetation growth, and they have been used to calculate other growth indices such as leaf area index (Keenan et al. 2014). The VIs calculated from PhenoCam imagery can also be used to record changes in the timing of phenophase transitions to detect how vegetation is responding to changes in local environment, such as changes brought on by climate change (Elmore et al. 2012; Killick et al. 2012; Ren et al. 2018). Four VIs that are prominent in phenology research include the green chromatic coordinate (GCC) (Richardson and Braswell 2009), the vegetation contrast index (VCI) (Zhang et al. 2018), the normalized difference vegetation index (NDVI) (Rouse et al. 1973) and the two-band enhanced vegetation index (EVI2) (Jiang et al. 2008).
The high temporal availability of PhenoCam imagery makes it a suitable data source for machine learning analysis. Also, the need for phenology models capable of detecting the underlying relationships between many environmental factors makes machine learning an important method to consider for the development of new models. The North American Great Plains provide an interesting study area to examine the interactions of different meteorological variables because of the spatial gradients that exist in temperature and precipitation. Because of this we sought to: (1) develop a regression model using XGB that can predict GCC, VCI, NDVI and EVI2 values using meteorological data at multiple grassland PhenoCam locations, (2) determine the primary meteorological variables within the model, and how these differ between VIs, and (3) predict the four VIs and measure their phenophases to establish trends in phenophase transitions using 38 years of historic meteorological data.
2 Methods and data
2.1 Study area
One of the 15 Level I ecoregions of North America, the Great Plains occupies 281 million ha with 224 million ha located within the contiguous U.S. (U.S. Environmental Protection Agency 2020). The Great Plains Ecoregion is divided into five Level II ecoregions: temperate prairies, west-central semiarid prairies, south-central semiarid prairies, Texas-Louisiana coastal plain and Tamaulipas-Texas semiarid plain (Fig. 1). We focused on the temperate prairies and the south-central semiarid prairies. Temperate prairies in the east are wetter and contain more croplands than the drier west-central and south-central semiarid prairies, while the west-central semiarid prairies are on average cooler than south-central semiarid prairies (Omernik and Griffith 2014).
The PhenoCam locations within the study area. Showing the six PhenoCam locations situated within the Great Plains of the contiguous U.S. Figure taken from Burke and Rundquist (2021)
We selected six grassland locations within the Great Plains (Fig. 1) each of which has a PhenoCam with at least three years of data (Table 1). Three of the sites are located within the temperate prairie ecoregion; the Oakville Prairie (Oakville), a part of the University of North Dakota, located in Grand Forks County, North Dakota (47.8993°N, 97.3161°W); the USGSEROS station at the Earth Resources Observation and Science (EROS) Data Center in South Dakota (43.7343°N, 96.6234°W); and the Nine Mile Prairie station (Nine-Mile), a part of the University of Nebraska – Lincoln (40.8680°N, 96.8221°W), located in Lancaster County, Nebraska. The other three PhenoCam sites are within the south-central semiarid prairie and are a part of the National Ecological Observatory Network (NEON). These sites include the NEON.D06.KONZ.DP1.00033 station (Konza) (39.1008°N, 96.5631°W) located at the Konza Prairie Biological Station in Kansas; the NEON.D10.ARIK.DP1.20002 station (ARIK) (39.7582°N, 102.4471°W) located near the Arikaree River in Yuma County, Colorado; and the NEON.D11.OAES.DP1.00033 station (OAES) (35.4106°N, 99.0588°W) located at the Klemme Range Research Station in Washita County, Oklahoma.
The six sites form a 1,470-km latitudinal transect through the Great Plains Ecoregion ranging from 35.4°N to 47.9°N. Oakville is part of the Level III/IV Ecoregion Lake Agassiz Plain/Saline Area, defined in part as having elevations between 250 and 265 MAMSL, annual precipitation ranging between 46 and 53 mm, and mean annual minimum/maximum temperatures between − 22°/−11 °C in January and 13°/28°C in July. EROS and Nine-Mile Prairie are in the Level III Western Corn Belt Plains. EROS is found within the Level IV Ecoregion Loess Prairies and Nine-Mile Prairie in the Glacial Drift Hills. Loess Prairie is characterized as having a range in elevation of 366 to 518 MAMSL with a mean annual precipitation between 58 and 64 mm and mean annual minimum/maximum temperatures ranging from − 13°/−1 °C in January to 17°/31°C in July (Bryce et al. 1996). The Glacial Drift Hills sit between 305 and 488 MAMSL with a mean annual precipitation between 69 and 89 mm and mean annual minimum/maximum temperatures between − 10°/1°C in January and 19°/33°C in July (Chapman et al. 2001).
The remaining three PhenoCams are in the Level II south-central semiarid prairies. Konza is located in the Level III Flint Hills, ranging from 305 to 488 MAMSL. That ecoregion’s annual precipitation is 71 to 89 mm. with mean annual minimum/maximum temperatures of −6°/6°C in January and 20°/36°C in July (Chapman et al. 2001). ARIK is part of the Level III/IV High Plains/Moderate Relief Plains. This ecoregion is found between 1,097 to 1,981 MAMSL. Its annual precipitation is between 30 and 46 mm and it has a mean annual minimum/maximum temperature range of −10°/7°C in January and 16°/33°C in July (Chapman et al. 2006). Finally, OAES is in the Level III/IV Central Great Plains/Rolling Red Hills, ranging from 427 to 792 MAMSL. This ecoregion’s annual precipitation is between 66 and 76 mm and its minimum/maximum mean annual temperature range is −8°/7°C in January and 19°/36°C in July (Woods et al. 2005).
2.2 PhenoCam data source and calculating the VIs
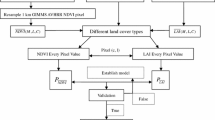
We choose to derive four VIs from the PhenoCam imagery at the six field stations. GCC (Eq. 1) is a proportional measure of relative ‘greenness’ that was originally developed for use with PhenoCams because of its relative stability under changing illumination conditions (Richardson and Braswell 2009). GCC has be used in a diverse array of ecosystem types, and can be measured using any digital capable of capturing a color (red, green, and blue) image (Richardson 2019). VCI (Eq. 2) was created as a nonlinear transformation of GCC that has a higher dynamic range relative to GCC by contrasting the green band to the sum of red and blue (Zhang et al. 2018). NDVI (Eq. 3) has a long history in Earth Observation (Rouse et al. 1973), and has been derived from PhenoCams that are sensitive to near-infrared wavelengths (Burke and Rundquist 2021; Filippa et al. 2018; Petach et al. 2014; Richardson 2019). EVI2 (Eq. 4) was developed as an adjustment to NDVI, with an enhanced ability to remove soil background noise, and atmospheric effects (Jiang et al. 2008).
To calculate each of the chosen VIs from the PhenoCam imagery, we first downloaded all available imagery from the six PhenoCam locations.Footnote 1 We then applied the exposure correction to both the color and mixed color-infrared imagery to extract the near-infrared and three color bands (Petach et al. 2014). Using the image digital numbers (DNs) for the red, green, blue (RGB) and near-infrared (NIR) bands the three VIs were calculated using Eqs. (1), (2), (3) and (4) for each day of the year in which PhenoCam imagery was available (Table 1). Finally, the PhenoCam VIs were linearly scaled to Gaussian Process Regression modeled VIs calculated with Harmonized Landsat-Sentinel surface reflectance imagery (described in detail in Burke and Rundquist 2021). This standardised the VI values between all PhenoCam sites, allowing them to be used together within a single XGB model.
2.3 Meteorological data
We used Daily Surface Weather and Climatological Summaries (DAYMET) data made available by the Oak Ridge National Laboratory (ORNL) within the Distributed Active Archive Center (DAAC) (Thornton et al. 2018). DAYMET provides 1 km x 1 km gridded data for North America starting in 1980, with several different weather variables available (Table 2). We retrieved the data for each of the six PhenoCam locations (Fig. 1), for the PhenoCam imagery time periods (Table 1).
We also used the DAYMET data to derive a few accumulative variables for precipitation, snow water equivalent (SWE) and temperature. Previous research has shown that precipitation often has a lag period before its has a measured effect on a VI’s signal (Potter and Brooks 1998; Wang et al. 2003). Based on this research we decided to accumulate precipitation over both 15 and 30 days to see if this would have a stronger relationship with the VI signals compared with the daily total precipitation. We did the same with the SWE, except changed the lag periods to 60 and 90 days to reflect the longer lag periods for snowfall. To calculate these values, we summed together the precipitation or SWE for the set number of days prior to each day of the year. To estimate the accumulated heat for vegetation growth we used growing degree days (GDD) calculated for each day of the year (Eq. 5) (Burke et al. 2018). GDD have historically been used for predicting agricultural crop growth and development, with Tbase set at 0 °C for winter wheat a C3 plant and 10 °C for corn a C4 plant (McMaster and Wilhelm 1997). We choose to calculate GDD for three Tbase values set at 0, 5 and 10 °C and examine the relationship these three datasets have with our grassland VIs. This resulted in a total of 13 variables being included in our model.
2.4 Statistical analysis of daily VIs
To produce a regression model for the four VIs we used XGB, a gradient boosted decision tree model (Chen and Guestrin 2016). We trained our XGB models using a randomly selected 80% (n = 2,815) of the available data, leaving 20% (n = 704) for model validation. To help prevent overfitting of the model, and to prune any branches with a negative gain, we set lambda to 1 and both alpha and gamma to 0. We also set the learning rate to 0.1, max depth to 10 and number of estimators to 50,000. We choose parameters that would help prevent overfitting of the model, and were recommended to produce a more conservative algorithm (Chen and Guestrin 2016). Subsampling, also know as bootstrap aggregating, was used so that a random selection of half (subsample = 0.5) the training samples were used to grow each tree with gradient-based selection (Chen and Guestrin 2016; Zhang et al. 2019).
Using the XBG model we fit each of the VIs against all the meteorological data variables including the accumulated precipitation, accumulated SWE and GDD. We combined the data sets across all six PhenoCam sites and created a model that could predict the four PhenoCam-based VIs at any one of the grassland sites given the daily meteorological data. By examining the total gain, a relative measure of a variable’s contribution to the model, we refined each of the VIs models further by removing the variables with the lowest total gain in a stepwise fashion until the R2 declined by more than 3% from the first model containing all variables, then selecting the model directly before the 3% decline. We used 3% as a threshold to minimize loss of model performance, while allowing enough of a reduction to the model to remove the variables that added little prediction power. Using the refined models for each of the four VIs we used the meteorological data to predict the VI values for each day of the year starting in 1981 and ending in 2019, producing a dataset for each VI ranging 38 years for each of the six PhenoCam locations.
2.5 Determining phenophase transitions dates
Using the 38 years of data for the four modeled VIs at the six PhenoCam locations we identified phenophase transitions dates using the same methods applied to the Collection 6 Moderate Resolution Imaging Spectrometer (MODIS) Land Cover Dynamics Product (CMCD12Q2) (Gray et al. 2019). The CMCD12Q2 product identifies seven phenophase stages throughout a growth cycle (Fig. 2), starting with greenup in the spring and ending with dormancy in the fall. This procedure was completed 24 times to account for the four VIs at 6 different sites. A natural cubic spline (Drury 2020) was fit to the full 38-year time series. To find the optimal number of knots to fit the spline we used Akaike’s Information Criterion (AIC) to balance under-overfitting of the model (Hurvich et al. 1998). To do this we randomly set aside one third of the dataset and fit the spline starting at 38 knots (1 knot per year of data) and ending at 570 knots (15 knots per year of data). Using the AIC we measured the models fit against the randomly removed data and selected the number of knots that produced the lowest AIC value. The spline was then re-fit to the entire dataset using the optimal number of knots.
Valid vegetation cycles were identified from the 24 spline models using methods similar to the CMCD12Q2 product (Gray et al. 2019). Local minima and maxima were identified for each year with a half year overlap at the beginning and end of the year. The maxima were examined for validity as a peak in vegetation growth while the minima were examined to be either the start or end of a vegetation cycle. However, the methods used for the CMCD12Q2 product were produced for EVI2 specifically. An amplitude change of 0.1 was required during any greenup or greendown period for it to be considered a valid cycle. The three other VIs have a varying range of values that do not necessarily align with EVI2. That is, instead of using a constant value of 0.1, we modified this step by requiring greenup and greendown periods to have an amplitude that is at least 70% that of the current year’s amplitude. Once the valid growth periods were identified we extracted the seven phenophase periods using the same methods as the CMCD12Q2 product. The peak is reached at the maximum value for the VI. The greenup, mid-greenup, and maturity occur at a 15, 50, and 90% increase in amplitude, while senescence, mid-greendown, and dormancy occur after the peak as amplitude decreases past 90, 50, and then 15%. Using these values, we also measured the length of greenup, the number of days between greenup and maturity, the length of maturity, the number of days between maturity and senescence, and the length of greendown, the number of days between senescence and dormancy, and the length of season, the number of days between greenup and dormancy.
3 Results
3.1 XGB regression models
Using the GCC, VCI, NDVI, and EVI2 datasets we produced four XGB regression models capable of predicting the VIs value based on all variables within the meteorological DAYMET data (Fig. 3). For each of the VIs 2,815 data points were used in model training, while 704 data points were set aside for model validation (Fig. 3). Examining the validation results GCC was the best fitting model with an R2 of 0.946 and a root mean square error (RMSE) of 0.01, while EVI2 was the poorest with an R2 of 0.895, and an RMSE of 0.02. Examining the total gain for each of the variables in the four models provides a relative measure of importance. Across all four models the photoperiod as day length, and temperature as GDD with a base of 0 °C were the two most important variables. While the minimum temperature and 30-days of accumulated precipitation were the third and fourth most important variables (Fig. 4). These four variables had the highest total gain across all four VIs, however they did not all occur in the same order. For example, day length had the highest total gain for GCC and VCI while GDD with a base of 0 °C was the highest for NDVI and EVI2.
3.2 Reducing the XGB regression models
With each of the four XGB regression models we removed variables one at a time for each VI independently, starting with the variable with the lowest total gain. We then refit the XGB models and assessed them with the validation dataset. We continued to remove variables until the R2 value of the validation dataset decreased by greater than 3% from the XGB models that contained all 13 meteorological variables, then selected the previous model. For the GCC and VCI XGB models this resulted in a final model using only four variables: day length, GDD with a base of 0 °C, 30-days of accumulated precipitation, and GDD with a base of 10 °C (Fig. 5). For the NDVI and EVI2 XGB models the final model required five variables: GDD with a base of 0 °C, day length, daily minimum temperature, 30-day accumulated precipitation, and GDD with a base of 5 °C (Fig. 5). These four XGB models were able to account for between 89.6 and 93.1% of the variation in the VIs datasets given 6 of the 13 meteorological variables (Fig. 6).
Using the four reduced VIs XGB regression models we conducted a sensitivity analysis to determine how a change in any of the variables effects the resulting VI value (Fig. 7). To do this we calculated the minimum, maximum and mean values for each of our variables, and then predicted the VI value at 100 evenly spaced sample points between each variable’s minimum and maximum while holding all other variables at their mean value. This analysis shows many of the nonlinearities between the meteorological variables and the VIs. For example, across all four VIs an increase in the lower values ( < ~ 1,000) of GDD 0 °C tends to cause an increase in the VI value. However, as GDD 0 °C increases ( > ~ 1,000), eventually the VI value either reaches a plateau or the VI starts decreasing as GDD 0 °C increases.
3.3 Trends in phenophase transitions
Using the XGB models with the 38 years of meteorological data we predicted the four VIs values for each day of the year. Then using these predictions splines were fit for the four VIs across the six PhenoCam locations. For example, at the Oakville station a spline model was fit to the predicted NDVI values (Fig. 8). Comparing the XGB predicted values with the spline models, we found that the splines were able to align well with an R2 and a RMSE ranging from 0.83 to 0.017 for GCC to 0.92 and 0.039 for NDVI (Fig. 9). Noticeably, the spline did reduce extreme values within the predicted VI values, for example in GCC where XGB predicted values below 0.2 were closer to 0.3 in the spline models. We examined the quantile range for both the XGB models and spline models and found little difference between the 1st, 2nd, and 3rd quantile for the two models, while the minimum and maximum values for the spline models were always closer to the median than the XGB models (Table 3).
For each of the spline models we predicted seven day of year (DOY) values as phenophases occurring within the vegetation growth cycles. We also calculated the length of greenup, the length of maturity, the length of greendown, and the total length of season, as the number of days between the greenup, maturity, senecence, and dormancy DOY values, respectively. This allowed us to examine trends in the seven phenophases to determine if over the 38-year data period they are occurring earlier of later in the growth cycle, and to determine if the lengths of time between them is increasing or decreasing. We calculated 66 linear regressions (Appendix 1–11), one for each phenophase (Appendix 1–7) and length (Appendix 8–11) between them at the six PhenoCam locations. Of these linear regressions we found 14 to have a significant trend within a 90% confidence interval (Table 4). The slope of these linear models provides us the change per year in each of the phenophases. For example, at the Oakville PhenoCam the dormancy phenophase produced a slope of 0.27, suggesting that dormancy is occurring 0.27 days later every year, which across our 38 years of data results in dormancy occurring 10 days later in 2019 compared to 1981.
4 Discussion
Using the XGB regression we developed a model capable of explaining 90 to 93% of the variability in four VIs (Fig. 6) across six grassland PhenoCam sites over the growing season. Our models demonstrate the importance of including photoperiod, temperature, and precipitation information when modeling vegetation phenology. Piao et al. (2019) reviewed the importance of including these different meteorological driving factors for modeling vegetation phenology and remarked that many current phenology models underperform because of their dependence on temperature without considering the interactions of other weather variables. A study by Wang et al. (2003) examined the Konza prairie, one of our six PhenoCam sites, and found that temperature was highly correlated with NDVI at the beginning and end of the growing season. Of the three GDD Tbase values explored, 0 °C remained the most import variable within our model, having the highest total gain and remaining in all four reduced models. A Tbase of 0 °C typically represents vegetation that uses the C3-pathway for photosynthesis such as grasslands in the temperate prairie region, while the C4-pathway is represented by a Tbase of 10 °C and would be more common in the hotter and drier south-central semiarid prairie (Jones and Vaughan 2010; McMaster and Wilhelm 1997). Because of this we anticipated that either the 0 °C and the 10 °C GDD variables would both be included in the reduced model or the 5 °C variable would better represent both regions and would have the highest total gain within the XGB regression. Instead, we found a mix of the three GDD Tbase values used depending on the VI (Fig. 5). Both reduced GCC and VCI models contained Tbase values 0 and 10 °C, while the NDVI and EVI2 contained Tbase values 0 °C, and 5 °C.
The stepwise backwards elimination in XGB regression model variables we used to refine our final model was a simple approach to limiting regression variables, while allowing the model to identify the most important variables to include. XGB models developed with 50 to hundreds of independent variables can use more advanced feature selection models eliminating multiple features at a time with optimization algorithms that speed up processing time (Pan et al. 2009; Zhang et al. 2019). With our approach, we were able to reduce our model from 13 variables down to four or five, depending on the VI, with a negligible change in model performance reflected in the average model R2 decreasing by 0.011 and RMSE increasing by 0.002. This reduction in model variables allowed us to examine the importance of the variables as well as the calculated lag times for precipitation and SWE, and the relationship between different Tbase values for GDD. Wang et al. (2003) found a two-week lag in NDVI’s response to precipitation events, however they also note that the response varied based on environmental conditions. For example, during a drier period the response to precipitation would often happen quicker. Our reduced models all selected precipitation with an accumulation of 30 days to best predict the phenology signals, suggesting that precipitation events occurring up to 30 days prior can control vegetation growth. This may be particularly true for the three PhenoCam sites in the south-central semiarid prairies since they are more susceptible to drought.
The four VIs we used across our analysis, GCC, VCI, NDVI, and EVI2, are all measures of vegetation phenology across the growing season. Of the three VIs, NDVI has the longest history in remote sensing (Rouse et al. 1973), while GCC has been well recognized within the PhenoCam literature because of its stability with uncalibrated imaging sensors (Richardson and Braswell 2009). VCI provides a nonlinear transformation of GCC, providing a higher range of values by contrasting green with the sum of red and blue (Zhang et al. 2018). EVI2 has also increased in use recently (Bolton et al. 2020; Peng et al. 2021), particularly with remotely sensed data from the Visible Infrared Imaging Radiometer Suite (VIIRS) system that lacks the blue band (Zhang et al. 2018).
Using the four VIs we were able to construct a 38-year phenology record at each PhenoCam location using the meteorological data and the reduced XGB models. Being able to use a combination of near-surface remote sensing and meteorological data to derive these VIs provides a valuable dataset for validation of satellite-based phenology products. It should be noted that these models reflect the vegetation from the period in which they were trained, 2015 to 2019. Any change in vegetation composition that may have occurred between 1981 and 2015 cannot be accounted for since this period of the models is based entirely on meteorological data and not on imagery from the PhenoCam stations. While this is a limitation of our models, it also acts as a control on our results since the trends in phenophase transition identified by the models are not affected by a change in species composition and are instead driven entirely by changes in climate. Changes in species composition can have a large effect on a phenology signal and presents a challenge in identifying climate change driven modification of phenophase transition periods (Prevéy and Seastedt 2014; Wilsey et al. 2018). Because our models are not based on imagery of the vegetation across the 38 years, and instead depend on meteorological data, we are able to model the timeseries under the assumption that the species composition did not change.
The spline models used for detecting the phenophase transitions were on average able to account for 87% of the variation in the models with RMSE ranging from 0.017 for GCC to 0.041 for VCI. One feature of the spline models we did note, was their tendency to be less influenced by extreme VI values (Table 3). Using the four splines for each VI at the six PhenoCam locations we measured seven phenophases and four phenophase periods. This resulted in 66 linear regression models (Appendix 1–11) to determine if any trends appeared in phenophase transitions over the 38-year timeseries. Examining the significant trends within a 90% confidence interval (Table 4) we found 14 phenophases that have shifted across the PhenoCam sites except for the Nine-Mile station, which had no significant trends. For the two northern PhenoCams in the temperate prairies the length of greendown has increased by 9.2 days (0.24 days/year) at the Oakville station, and 19.2 days (0.51 days/year) at the EROS station over the 38 years. The 10-day difference between the two stations is likely attributed to the fact that the EROS station has seen an earlier onset of peak greenness by 13.1 days (-0.35 days/year), and an earlier onset of senescence by 11.7 days (-0.31 days/year), which has also shortened the length of maturity by 7.4 days (-0.19 days/year). This suggests that the growing season at the EROS station is trending towards a quicker occurrence of peak greenness followed by a shorter period of greenness between maturity and senescence, with an extension in the greendown period. In a study using imagery from the Advanced Very High Resolution Radiometer (AVHRR) from 1982 to 2002, Reed (2006) found grasslands to have a later dormancy period by 6.52 days (0.33 days/year), while greenup also started later by 8.01 days (0.40 days/year). A similar study used AVHRR from 1982 to 2006, Zhu et al. (2012) found grasslands in North America to have a later onset of greenness by 7.6 days (0.32 days/year), and a later dormancy by 2.1 days (0.09 days/year) causing a shortening of the growing season by 5.6 days (-0.23 days/year). The offset of dormancy occurring later into the season agrees with our study with dormancy at the Oakville station occurring 10 days later (0.27 days/year). This falls within the range found by Liu et al. (2016) with dormancy in the Northern Hemisphere occurring between 0.19 and 0.45 days later each year. For five of the six PhenoCam sites greenup did not have a significant trend, with no sites finding greenup occurring later. The one site with a greenup trend was the Konza station in which greenup occurs 9.5 days (-0.25 days/year) earlier in 2019 then in 1981. This value is close to the 2.8 days per decade (-0.28 days/year) in which spring phenology is predicted to have advanced for both plants and animals in the northern hemisphere (Hoegh-Guldberg et al. 2018). At the Konza station maturity and peak greenness is also occurring earlier in the year by 10.4 days (-0.27 days/year) and 11.5 days (-0.30 days/year), respectively. For this station, the earlier onset of greenness seems to be to be followed by an earlier onset of maturity and peak greenness for the vegetation. Of the six stations ARIK was the only station to find a significant trend in the overall length of the growing season with it increasing by 23.3 days (0.61 days/year). This station also had its length of greenup increase by 16.2 days (0.43 days/year) while its senescence and mid-greendown dates are occurring 7.5 days (0.20 days/year) and 13.7 days (0.36 days/year) later, respectively. The ARIK increase in length of season agrees with Zhou et al. (2001) who used AVHRR from 1981 to 1999 finding length of season in North America to increase on average by 12 days (0.65 days/year) and finding dormancy to occur 4 days (0.22 days/year) later. Overall across the five PhenoCam locations the significant trends we found align with studies of vegetation phenology over North American grasslands. Jeong et al. (2011) used AVHRR to assess phenology from 1982 to 2008 and found both temporal and spatial variations in different phenology trends. They identified a reduction in the trend of an earlier onset greenness starting in 2000, while at the same time found an increased rate in later onset of dormancy, with both contributing to a lengthening of the growing season. While we did have variability in growing season length across our six field stations, as expected with increased latitudinal variability in Spring temperatures for North American grasslands that can have an influence on both spring and fall phenology (Liu and Zhang 2020). Across our study area the results indicate that changing temperature and precipitation patterns are driving a significant change in phenology of the grasslands.
5 Conclusion
We used the machine learning based XGB regression model to predict changes in GCC, VCI, NDVI, and EVI2 across the growing season at six PhenoCam sites. With this model we were able to accurately predict 90 to 93% of variability in the VI values. This allowed us to reconstruct the VIs signals to derive a 38-year timeseries. With these modeled timeseries we were able to examine the trending changes in the phenophases at each of the grassland field sites. In the temperate prairies, the length of greendown has increased by 9.2 days at Oakville and 19.2 days at EROS, while we did not find any significant shift in Nine-Mile’s phenophases. In the South-central semiarid prairies, Konza shows a trend in greenup, which occurs 9.5 days earlier in 2019 than in 1981; ARIK has a significant trend in the overall length of the growing season, increasing by 23.3 days, and we see a significant positive trend on the length of plant maturity at OAES. The significant trends we identified agreed with the many AVHRR and other satellite-based analysis that have been done for North American grasslands. We believe the methods used to develop our framework provide a valuable framework for future work modeling vegetation phenology. Using near-surface remote sensing and meteorological data provides a valuable validation dataset for satellite-based phenology. Our model can be applied to additional PhenoCam sites, including ecosystem types other than grasslands, to examine the interactions between photoperiod, temperature, and precipitation in these regions. Also, additional environmental factors could be considered such as soil moisture or nutrient availability. Future work that would help improve our understanding of grassland phenology should focus on identifying the spatial and temporal variability that exists in the phenology of the North American Great Plains. In addition, our framework should be tested with data gathered by other Earth observation sensors and in other geographic regions.
Data availability
The dataset analyzed during the current study is not publicly available.
References
Anderson JM (1991) The effects of Climate Change on decomposition processes in Grassland and Coniferous forests. Ecol Appl 1:326–347
Belda S, Pipia L, Morcillo-Pallarés P, Rivera-Caicedo JP, Amin E, De Grave C, Verrelst J (2020) DATimeS: a machine learning time series GUI toolbox for gap-filling and vegetation phenology trends detection. Environ Model Softw 127. https://doi.org/10.1016/j.envsoft.2020.104666
Bolton DK, Gray JM, Melaas EK, Moon M, Eklundh L, Friedl MA (2020) Continental-scale land surface phenology from harmonized landsat 8 and Sentinel-2 imagery. Remote Sens Environ 240:111685. https://doi.org/10.1016/j.rse.2020.111685
Browning DM, Karl JW, Morin D, Richardson AD, Tweedie CE (2017) Phenocams bridge the gap between field and satellite observations in an arid grassland ecosystem. Remote Sens 9:10. https://doi.org/10.3390/rs9101071
Bryce SA, Omernik JM, Pater DA, Ulmer M, Schaar J, Freeouf J, Johnson R, Kuck P, Azevedo SH (1996) Ecoregions of North Dakota and South Dakota, (color poster with map, descriptive text, summary tables, and photographs): Reston, Virginia, U.S. Geological Survey (map scale 1:1,500,000)
Burke MWV, Rundquist BC (2021) Scaling Phenocam GCC, NDVI, and EVI2 with Harmonized Landsat-Sentinel using gaussian processes. Agric for Meteorol 300:108316. https://doi.org/10.1016/j.agrformet.2020.108316
Burke MWV, Shahabi M, Xu Y, Zheng H, Zhang X, Vanlooy J (2018) Identifying the driving factors of water quality in a sub-watershed of the republican river basin, Kansas USA. Int J Environ Res Public Health 15:5. https://doi.org/10.3390/ijerph15051041
Chapman SS, Omernik JM, Freeouf JA, Huggins DG, McCauley JR, Freeman CC, Steinauer G, Angelo RT, Schlepp RL (2001) Ecoregions of Nebraska and Kansas (color poster with map, descriptive text, summary tables, and photographs): Reston, Virginia, U.S. Geological Survey (map scale 1:1,950,000)
Chapman SS, Griffith GE, Omernik JM, Price AB, Freeouf J, Schrupp DL (2006) Ecoregions of Colorado (color poster with map, descriptive text, summary tables, and photographs): Reston, Virginia, U.S. Geological Survey (map scale 1:1,200,000)
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. Proc ACM SIGKDD Int Conf Knowl Discovery Data Min 13–17–Augu:785–794. https://doi.org/10.1145/2939672.2939785
Cleland EE, Chiariello NR, Loarie SR, Mooney HA, Field CB (2006) Diverse responses of phenology to global changes in a grassland ecosystem. Proceedings of the National Academy of Sciences 103:13740–13744. https://doi.org/10.1073/pnas.0600815103
Dai W, Jin H, Zhang Y, Liu T, Zhou Z (2019) Detecting temporal changes in the temperature sensitivity of spring phenology with global warming: application of machine learning in phenological model. Agric for Meteorol 279:107702. https://doi.org/10.1016/j.agrformet.2019.107702
Drury M (2020) Basis Expansions. https://github.com/madrury/basis-expansions
Elmore AJ, Guinn SM, Minsley BJ, Richardson AD (2012) Landscape controls on the timing of spring, autumn, and growing season length in mid-atlantic forests. Glob Change Biol 18:656–674. https://doi.org/10.1111/j.1365-2486.2011.02521.x
Filippa G, Cremonese E, Migliavacca M, Galvagno M, Sonnentag O, Humphreys E, Hufkens K, Ryu Y, Verfaillie J, di Morra U, Richardson AD (2018) NDVI derived from near-infrared-enabled digital cameras: Applicability across different plant functional types. Agric for Meteorol 249:275–285. https://doi.org/10.1016/j.agrformet.2017.11.003
Flanagan LB, Adkinson AC (2011) Interacting controls on productivity in a northern Great Plains grassland and implications for response to ENSO events. Glob Change Biol 17:3293–3311. https://doi.org/10.1111/j.1365-2486.2011.02461.x
Fu YH, Zhao H, Piao S, Peaucelle M, Peng S, Zhou G, Ciais P, Huang M, Menzel A, Peñuelas J, Song Y, Vitasse Y, Zeng Z, Janssens IA (2015) Declining global warming effects on the phenology of spring leaf unfolding. Nature 526:104–107. https://doi.org/10.1038/nature15402
Gray J, Sulla-Menashe D, Friedl MA (2019) User Guide to Collection 6 MODIS Land Cover Dynamics (MCD12Q2) Product 6:1–8
Henebry GM (2013) Phenologies of North American Grasslands and Grasses. In Mark D. Schwartz (Ed.), Phenology: An Integrative Environmental Science (2nd ed.). Springer. https://doi.org/10.1007/978-94-007-6925-0_11
Hoegh-Guldberg O, Jacob D, Taylor M, Bindi M, Brown S, Camilloni I, Diedhiou A, Djalante R, Ebi KL, Engelbrecht F, Guiot J, Hijioka Y, Mehrotra S, Payne A, Seneviratne SI, Thomas A, Warren R, Zhou G (2018) Impacts of 1.5oC Global Warming on Natural and Human Systems. In Special Report, Intergovernmental Panel on Climate Change (Issue ISBN 978-92-9169-151-7)
Hufkens K, Keenan TF, Flanagan LB, Scott RL, Bernacchi CJ, Joo E, Brunsell NA, Verfaillie J, Richardson AD (2016) Productivity of North American grasslands is increased under future climate scenarios despite rising aridity. Nat Clim Change 6:710–714. https://doi.org/10.1038/nclimate2942
Hurvich CM, Simonoff JS, Tsai CL (1998) Smoothing parameter selection in nonparametric regression using an improved Akaike information criterion. J R Stat Soc Series B Stat Methodol 60:271–293. https://doi.org/10.1111/1467-9868.00125
Jeong SJ, Ho CH, Gim HJ, Brown ME (2011) Phenology shifts at start vs. end of growing season in temperate vegetation over the Northern Hemisphere for the period 1982–2008. Glob Change Biol 17:2385–2399. https://doi.org/10.1111/j.1365-2486.2011.02397.x
Jiang Z, Huete AR, Didan K, Miura T (2008) Development of a two-band enhanced vegetation index without a blue band. Remote Sens Environ 112:3833–3845. https://doi.org/10.1016/j.rse.2008.06.006
Jones HG, Vaughan RA (2010) Remote sensing of vegetation: principles, techniques, and applications, 1st edn. Oxford University Press
Keenan TF, Darby B, Felts E, Sonnentag O, Friedl MA, Hufkens K, O’Keefe J, Klosterman S, Munger JW, Toomey M, Richardson AD (2014) Tracking forest phenology and seasonal physiology using digital repeat photography: A critical assessment. Ecol. Appl. 24:1478–1489. https://doi.org/10.1890/13-0652.1
Killick R, Fearnhead P, Eckley IA (2012) Optimal detection of changepoints with a linear computational cost. J. Am. Stat. Assoc. 107:1590–1598. https://doi.org/10.1080/01621459.2012.737745
Liu L, Zhang X (2020) Effects of temperature variability and extremes on spring phenology across the contiguous United States from 1982 to 2016. Sci Rep 10:1–14. https://doi.org/10.1038/s41598-020-74804-4
Liu Q, Fu YH, Zhu Z, Liu Y, Liu Z, Huang M, Janssens IA, Piao S (2016) Delayed autumn phenology in the Northern Hemisphere is related to change in both climate and spring phenology. Glob Change Biol 22:3702–3711. https://doi.org/10.1111/gcb.13311
McMaster GS, Wilhelm WW (1997) Growing degree-days: one equation, two interpretations. Agric for Meteorol 87:291–300. https://doi.org/10.1016/S0168-1923(97)00027-0
Omernik JM, Griffith GE (2014) Ecoregions of the Conterminous United States: evolution of a hierarchical spatial Framework. Environ Manage 54:1249–1266. https://doi.org/10.1007/s00267-014-0364-1
Pan F, Converse T, Ahn D, Salvetti F, Donato G (2009) Feature selection for ranking using boosted trees. International Conference on Information and Knowledge Management, Proceedings 2025–2028. https://doi.org/10.1145/1645953.1646292
Peng D, Wang Y, Xian G, Huete AR, Huang W, Shen M, Wang F, Yu L, Liu L, Xie Q, Liu L, Zhang X (2021) Investigation of land surface phenology detections in shrublands using multiple scale satellite data. Remote Sensing of Environment 252. https://doi.org/10.1016/j.rse.2020.112133
Petach AR, Toomey M, Aubrecht DM, Richardson AD (2014) Monitoring vegetation phenology using an infrared-enabled security camera. Agric for Meteorol 195–196:143–151. https://doi.org/10.1016/j.agrformet.2014.05.008
Petrie MD, Brunsell NA, Vargas R, Collins SL, Flanagan LB, Hanan NP, Litvak ME, Suyker AE (2016) The sensitivity of carbon exchanges in Great Plains grasslands to precipitation variability. J Geophys Research: Biogeosciences 121:280–294. https://doi.org/10.1002/2015JG003205
Piao S, Liu Q, Chen A, Janssens IA, Fu Y, Dai J, Liu L, Lian X, Shen M, Zhu X (2019) Plant phenology and global climate change: current progresses and challenges. Glob Change Biol 14619. https://doi.org/10.1111/gcb.14619
Potter CS, Brooks V (1998) Global analysis of empirical relations between annual climate and seasonality of NDVI. Int J Remote Sens 19:2921–2948. https://doi.org/10.1080/014311698214352
Prevéy JS, Seastedt TR (2014) Seasonality of precipitation interacts with exotic species to alter composition and phenology of a semi-arid grassland. J Ecol 102:1549–1561. https://doi.org/10.1111/1365-2745.12320
Reed BC (2006) Trend analysis of time-series phenology of North America derived from satellite data. GIScience Remote Sens 43:24–38. https://doi.org/10.2747/1548-1603.43.1.24
Ren S, Chen X, Lang W, Schwartz MD (2018) Climatic controls of the spatial patterns of vegetation phenology in Midlatitude grasslands of the Northern Hemisphere. J Geophys Research: Biogeosciences 123:2323–2336. https://doi.org/10.1029/2018JG004616
Richardson AD (2019) Tracking seasonal rhythms of plants in diverse ecosystems with digital camera imagery. New Phytol 222:1742–1750. https://doi.org/10.1111/nph.15591
Richardson A, Braswell B (2009) Near-surface remote sensing of spatial and temporal variation in canopy phenology. Ecol Appl 19:417–1428. http://www.esajournals.org/doi/abs/https://doi.org/10.1890/08-2022.1
Richardson AD, Hufkens K, Milliman T, Aubrecht DM, Chen M, Gray JM, Johnston MR, Keenan TF, Klosterman ST, Kosmala M, Melaas EK, Friedl MA, Frolking S (2018) Tracking vegetation phenology across diverse north American biomes using PhenoCam imagery. Sci Data 5:180028. https://doi.org/10.1038/sdata.2018.28
Rouse JWJ, Haas RH, Schell JA, Deering DW (1973) Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation
Schwartz MD, Ahas R, Aasa A (2006) Onset of spring starting earlier across the Northern Hemisphere. Glob Change Biol 12:343–351. https://doi.org/10.1111/j.1365-2486.2005.01097.x
Scurlock JMO, Hall DO (1998) The global carbon sink: a grassland perspective. Glob Change Biol 4:229–233
Stubbendieck J, Hatch SL, Dunn CD (2017) Grasses of the Great Plains. First). Texas A&M University
Thornton PE, Thornton MM, Mayer BW, Wei Y, Devarakonda R, Vose RS, Cook RB (2018) Daymet: Daily Surface Weather Data on a 1-km Grid for North America, Version 3. In ORNL DAAC. https://doi.org/10.3334/ORNLDAAC/1328
U.S. Environmental Protection Agency (2020) Ecoregions of North America. https://www.epa.gov/eco-research/ecoregions-north-america
Wang J, Rich PM, Price KP (2003) Temporal responses of NDVI to precipitation and temperature in the central Great Plains, USA. Int J Remote Sens 24:2345–2364. https://doi.org/10.1080/01431160210154812
Wilsey BJ, Martin LM, Kaul AD (2018) Phenology differences between native and novel exotic-dominated grasslands rival the effects of climate change. J Appl Ecol 55:863–873. https://doi.org/10.1111/1365-2664.12971
Woods AJ, Omernik JM, Butler DR, Ford JG, Henley JE, Hoagland BW, Arndt DS, Moran BC (2005) Ecoregions of Oklahoma (color poster with map, descriptive text, summary tables, and photographs): Reston, Virginia, U.S. Geological Survey (map scale 1:1,250,000)
World Wildlife Fund (2018) The Plowprint Report: 2018. worldwildlife.org/ngp
Zhang X, Jayavelu S, Liu L, Friedl MA, Henebry GM, Liu Y, Schaaf CB, Richardson AD, Gray J (2018) Evaluation of land surface phenology from VIIRS data using time series of PhenoCam imagery. Agric for Meteorol 256–257:137–149. https://doi.org/10.1016/j.agrformet.2018.03.003
Zhang H, Eziz A, Xiao J, Tao S, Wang S, Tang Z, Zhu J, Fang J (2019) High-resolution vegetation mapping using eXtreme gradient boosting based on extensive features. Remote Sens 11:12. https://doi.org/10.3390/rs11121505
Zhou L, Tucker CJ, Kaufmann RK, Slayback D, Shabanov NV, Myneni RB (2001) Variations in northern vegetation activity inferred from satellite data of vegetation index during 1981 to 1999. J Geophys Res Atmos 106:20069–20083. https://doi.org/10.1029/2000JD000115
Zhu W, Tian H, Xu X, Pan Y, Chen G, Lin W (2012) Extension of the growing season due to delayed autumn over mid and high latitudes in North America during 1982–2006. Glob Ecol Biogeogr 21:260–271. https://doi.org/10.1111/j.1466-8238.2011.00675.x
Acknowledgements
We thank Earl Klug, Justine Burke, and Mbongowo Mbuh for helping with field data collection, and all the UND students who have helped maintain the Oakville Phenocam, as well as the UND Field Station Committee for funding the mobile data connection. We would also like to thank the Phenocam Network, and those who have maintained the field locations we used, as well as Jeffrey VanLooy, Haochi Zeng, Sean Hammond, Gregory Vandeberg, and Jesslyn Brown who served on the doctoral committee.
Funding
Authors Morgen Burke and Anaí Caparó Bellido received research support from the University in the form of Graduate Research Assistantships.
Author information
Authors and Affiliations
Contributions
The study was conceptualized and designed by MB and BR. Material preparation, data collection and analysis were performed by MB. The manuscript was written, read and approved by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Burke, M.W.V., Rundquist, B.C. & Caparó Bellido, A. Modelling vegetation phenology at six field stations within the U.S. Great Plains: constructing a 38-year timeseries of GCC, VCI, NDVI, and EVI2 using PhenoCam imagery and DAYMET meteorological records. Theor Appl Climatol 155, 5219–5235 (2024). https://doi.org/10.1007/s00704-024-04933-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00704-024-04933-7