Abstract
Plastid genomes have been widely applied to elucidate plant evolution at higher taxonomic levels, but have rarely been considered useful for addressing close relationships. Here, we resolve the phylogeny and taxonomy of the Chinese lianoid Gnetum clade (Gnetales), using high throughput and Sanger sequencing techniques and studies of plant morphology. Despite previous efforts, relationships among taxa and the taxonomy within the clade have remained unclear. We generated 11 plastid genomes representing one arborescent and four lianoid species. Phylogenetic analyses were conducted using (a) the entire plastid genomes and (b) the protein-coding genes only. Sequence divergence among the lianoid species was substantial, with 9345 variable sites. Four variable regions were identified, targeted and sequenced for an additional 54 specimens and analyzed together with one nuclear ribosomal marker. Results from the phylogenetic analyses corroborate G. parvifolium as sister to the remaining lianoid species and support the presence of at least five additional species in the Chinese lianoid clade: G. catasphaericum, G. formosum, G. luofuense, G. montanum and G. pendulum. Following morphological investigations, G. giganteum and G. gracilipes are included in and synonymized with G. pendulum. Gnetum hainanense is included in and synonymized with G. luofuense. Two names, G. indicum and G. cleistostachyum, remain questionable. A taxonomic revision and a key to Chinese lianoid Gnetum are presented. Internal nodes in the Chinese lianoid Gnetum clade are from the Miocene and onwards and coincide with the expansion of tropical to subtropical forests in South China, which may have facilitated speciation in the clade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, next-generation sequencing (NGS) has revolutionized systematic biology and molecular ecology. Compared to traditional Sanger sequencing techniques, NGS allows us to generate orders of magnitude of more informative characters in a both cost- and time-efficient way (Davey et al. 2011; Ekblom and Galindo 2011; Straub et al. 2012). As a consequence, NGS has allowed for the description of a rapidly increasing number of nuclear, mitochondrial and plastid genomes (Rounsley et al. 2009; Deschamps and Campbell 2010) and is set to contribute to a deeper understanding of both phylogeny and evolution in many lineages. Among the three types of genomes, data of entire plastid genomes have been widely used to seek for markers for the purpose of DNA barcoding (Hollingsworth et al. 2011; Nock et al. 2011), to investigate population genetics, and in phylogeographic studies (Powell et al. 1995; Provan et al. 2001; Mariac et al. 2014), as well as to efficiently address phylogenetic questions in both angiosperms (Jansen et al. 2007; Moore et al. 2011; Huang et al. 2014) and gymnosperms (Parks et al. 2009; Lin et al. 2010; Yi et al. 2015). Here, we use data from NGS, combined with data from traditional Sanger sequencing and morphology, with the primary aim of resolving the phylogeny and species delimitation among Chinese lianoid species of Gnetum L. (Gnetales).
Gnetum consists of about 30–40 species of trees, shrubs and lianas that inhabit lowland (sub-)tropical forests of South America, Africa and Asia (Kubitzki 1990; Price 1996; Hou et al. 2015). Most of the species diversity is found in the eastern and southeastern regions of Asia, where vegetative parts and seeds from Gnetum have been used for medical purposes (Yao and Lin 2005; Yao et al. 2006; Liu et al. 2010), for production of daily necessities (Markgraf 1951) and as food and drinks (Fu et al. 1999a). The genus is morphologically characterized by opposite and decussate phyllotaxy, simple and petiolate leaves with entire leaf margins, pinnate and reticulate venation (Fig. 1a, b) and a dioecious habit where fertile spikes bear several collars on which whorls of male and/or female reproductive units attach (Fig. 1c, d male spikes, e–g female spikes). Sterile female units are typically present in male spikes, where they can be exposed or hidden among hairs and the male units of the collars. Seeds of Gnetum have outer fleshy coats and an inner stony layer (Fig. 1g–p) produced from extraovular seed envelopes. Depending on species, seeds are either sessile (Fig. 1i, j) or stipitate (Fig. 1k–p).
Vegetative and reproductive morphology of Chinese Gnetum. a Leaves and branches of G. parvifolium. b Leaves of G. luofuense. c Male spikes of G. montanum. d A male spike of G. luofuense (=G. hainanense). e Female spikes of G. luofuense (=G. hainanense). f A female spike of G. catasphaericum. g A female spike of G. parvifolium with a developing seed. h Seeds of G. luofuense (=G. hainanense) in clusters. Seeds of Chinese lianoid Gnetum. i G. parvifolium. j G. formosum. k G. catasphaericum. l G. montanum. m–n G. pendulum. o, p G. luofuense (=G. hainanense).
The present study focuses on Chinese lianoid species of Gnetum (clade O sensu, Hou et al. 2015). Markgraf, who monographed the genus in 1930, recognized one arborescent taxon (G. gnemon L.) and two lianoid taxa from China (G. montanum Markgr. and G. montanum f. parvifolium (Warb.) Markgr.). Later, several new taxa were described (Markgraf 1930; Cheng 1964; Cheng et al. 1975; Shao 1994; Fu et al. 1999b), and at least 11 Chinese lianoid taxa have been proposed and discussed in the past. However, the taxonomic identity and delimitation of some of these taxa are unclear, partly as a result of insufficient knowledge of their genotypic and phenotypic variation, but also as a result of formal problems in their original protologues. Markgraf (1930), for example, listed no less than 43 syntypes in the original protologue of G. montanum, a heterogeneous collection that probably includes representatives of more than one species (Fu et al. 1999a). Other names (taxa), such as G. cleistostachyum C.Y.Cheng, G. pendulum f. intermedium C.Y.Cheng, were incorrectly described. Consequently, the names are formally invalid, and if they truly point at taxonomically identifiable units (species), they should be validly published and described. Relationships among the lianoid taxa are also unclear. Previous phylogenetic studies based on molecular data support that two arborescent species, G. gnemon and G. costatum K.Schum., comprise the sister clade to the remaining Asian (lianoid) Gnetum (Hou et al. 2015). Among the Asian lianoid Gnetum, Chinese species constitute a strongly supported clade (Won and Renner 2003, 2005a, 2006; Hou et al. 2015), but relationships within this clade remain poorly supported, partly due to restricted sampling in previous phylogenetic analyses, but also because of limited sequence variation in the utilized molecular markers. Another problem may be misidentified specimens, resulting in contradictory taxonomic conclusions in the past.
Here, we aim to resolve the relationships and taxonomy of the Chinese lianoid clade of Gnetum using a combination of molecular and morphological data. We have developed new markers with appropriate amount of variation by generating complete plastid genomes of representative species. Developed markers are subsequently sequenced for additional specimens, and relationships of the included specimens are inferred by conducting phylogenetic analyses using all generated sequence data. Results, in combination with morphological investigations, are used to revise the taxonomy of the group. We also assess node ages in the clade and discuss the underlying evolutionary history of Chinese lianoid Gnetum.
Materials and methods
Taxon sampling
Silica material for DNA extraction was collected during field trips to the southern provinces of China (Guangdong, Guangxi, Yunnan and Hong Kong). In addition, material from herbaria in Belgium (BR), Scotland (E), China (HITBC, PE), Indonesia (KRB), USA (MO), Sweden (S) and the Netherlands (U) were studied. Altogether representatives of nine lianoid species (G. catasphaericum H.Shao, G. formosum Markgr., G. giganteum H.Shao, G. gracilipes C.Y.Cheng, G. hainanense C.Y.Cheng ex L.K.Fu, Y.F.Yu & M.G.Gilbert, G. luofuense C.Y.Cheng, G. parvifolium (Warb.) W.C.Cheng, G. pendulum and G. montanum) were included in the phylogenetic analyses (Online Resource 1), among which G. catasphaericum, G. formosum, G. giganteum, G. gracilipes and G. pendulum had previously never been included in such analyses. Eleven specimens, representing four lianoid species (G. hainanense, G. parvifolium, G. pendulum and G. montanum) and one arborescent species (G. gnemon), were used for generating NGS data and complete plastid assembly. Subsequent analyses, also using Sanger sequencing data, included a total of 66 specimens, 50 representing nine Chinese lianoid species, 15 representing remaining Asian taxa and one specimen representing the African species G. africanum Welw. Specimens representing G. cleistostachyum and G. indicum were not available for molecular studies.
Plastid genome assembly, annotation and alignment
Total genomic DNA of Gnetum was extracted from silica-treated leaves according to the cetyltrimethylammonium bromide (CTAB) method (Doyle 1991) and purified using a QIAquick PCR Purification Kit (Qiagen, Sweden). To comply with the DNA quantity sample requirement of the Illumina platform, silica material of each specimen was sampled twice and manually merged for subsequent DNA library construction. High-throughput sequencing was performed at Science for Life Laboratory (Stockholm, Sweden) following the manufacturer’s instructions for the Illumina HiSeq 2500 platform (Illumina, San Diego, USA). Single raw reads from Illumina sequencing were set in pairs, and low-quality nucleotides were removed with the error probability limit 0.05 in Geneious version 8.0.5 (Kearse et al. 2012). Trimmed paired reads were mapped onto the plastid genome of G. parvifolium (GenBank ID, NC_011942) as the reference for five iterations with medium to low sensitivity in the Geneious mapper (installed in the same software). Eleven draft sequences of plastid genomes were generated, and matched raw reads for each specimen were saved. De novo assembly of matched raw reads was performed for each specimen using assembler Velvet version 1.2.10 (Zerbino and Birney 2008) with the K-mer value optimized to 99 using Velvet Optimizer (installed in the same software). To evaluate accuracy, generated de novo contigs of each specimen were aligned with the draft plastid genomes from mapping. The quality of draft genomes were assessed and manually adjusted to fill up gaps and unclear nucleotides where necessary. Complete plastid genome sequences were annotated using DOGMA (Wyman et al. 2004) with the default settings. The start and stop codons of protein-coding genes were inferred by comparison with the known plastid genomes of the Gnetales. Annotated tRNA was further assessed using tRNAscan-SE version 1.21 (Schattner et al. 2005). A total of 11 complete plastid genomes of Gnetum have been deposited in GenBank (ID: KX385188–KX385198, Online Resource 1). A multiple alignment of the 11 genomes was performed using MAFFT version 7.017 (Katoh et al. 2002) with the following settings: scoring matrix 200PAM/k = 2, algorithm setting FFT-NS-2. Sequences of protein-coding regions were manually modified in the multiple alignments, and aligned plastid genomes with annotations were visualized using mVISTA (Mayor et al. 2000) by comparison with the reference plastid genome.
Pairwise genome comparisons, marker designing and PCR amplification
To detect divergence hotspots in the plastid genome, the alignment of 11 complete plastid genomes was skimmed using Geneious variant/SNP finder with the minimum variant frequency set to 0.1 and maximum variant P value set to 10−6. Divergence hotspots consist of nucleotide substitutions and indels (insertions and deletions), which are marked onto the consensus of the alignment. With regard to sequence variations revealed in mVISTA and detected using variants/SNPs finder, four plastid markers, i.e., matK, rpoC1, psbB-rps12 and trnF-trnV, were chosen to address species relationships of Chinese lianoid Gnetum. We made initial attempts to amplify three additional markers (ycf1, trnP-ycf1, trnK-trnT) but encountered problems and these markers were not further used. It is probably due to large numbers of sequence repeats in these regions of the plastid genomes of Gnetum. Complementing the four plastid markers, also the nuclear ribosomal marker ITS1 (including partial 5.8S) was utilized, a marker that has been used to resolve species relationships in Gnetum previously (e.g., Won and Renner 2005b). All primer information is listed in Table 1. PCR conditions for nrITS regions were set to an initial denaturation at 97 °C for 1 min; followed by 40 cycles of 97 °C for 10 s, 55 °C for 30 s and 72 °C for 20 s (4 s were added at each cycle); and followed by a final extension at 72 °C for 7 min. For amplification of plastid markers, the initial denaturation was set to 95 °C for 2 min; followed by 37 cycles of 95 °C for 30 s, 50 °C for 45 s and 72 °C for 2 min; and followed by a final extension at 72 °C for 7 min. PCR products were purified using Multiscreen PCR purification kits (Millipore, Billerica, MA) and sequenced at Macrogen Europe (Netherland). Sequences from Sanger sequencing were assembled using the Staden Package version 1.6 (Staden 1996) and aligned using Clustal W version 1.81 (Thompson et al. 1994). Multiple alignments for each marker were visualized and manually adjusted in Mesquite version 2.75 (Maddison and Maddison 2011).
Phylogenetic analyses of NGS data
Phylogenetic analyses of the NGS data were conducted using two datasets: dataset (1) comprising the 11 complete plastid genomes, and dataset (2) comprising a concatenation of all the protein-coding genes from the 11 plastid genomes (Table 2, Online Resource 2). Data in each dataset were treated as a single partition and analyzed using both maximum likelihood and a Bayesian inference approach. The single partition was tested with 22 candidate substitution models using jModeltest version 2.1.3 (Darriba et al. 2012). Best-fitting model was selected according to the Akaike information criterion (AIC), and a general time reversible substitution model with substitution rates drawn from a gamma distribution (GTR + Γ) was selected. Maximum likelihood analyses were conducted using GTR + Γ model implemented in RAxML version 7.2.8 (Stamatakis 2014). The analyses were conducted with 1000 replicates of rapid bootstrapping using a parsimony tree as a starting tree. Two specimens of Gnetum gnemon were used as outgroup, based on the results in previous phylogenetic studies (Won and Renner 2006; Hou et al. 2015). Bayesian analyses were conducted using MrBayes version 3.2.3 (Ronquist et al. 2012) via the online service of Cyber-infrastructure for Phylogenetic Research (CIPRES, Miller et al. 2010). Two parallel runs were set, each with four chains and using GTR + Γ as substitution model. The analyses were run for six million generations with a sampling frequency of 1000 generations. Convergence of the two MCMC runs was assessed using software AWTY (Nylander et al. 2008) and Tracer version 1.6 (Rambaut and Drummond 2003). Sampled trees were summarized in a majority rule consensus with the initial 600 trees removed as burn-in.
Phylogenetic analyses of Sanger sequencing data
Phylogenetic analyses were also conducted on a 66-taxon dataset generated from Sanger sequencing. Two different datasets were applied: dataset 3) a two-partitioned dataset of nrITS and the combined four plastid markers (matK, rpoC1, psbB-rps12 and trnF-trnV) and dataset 4) a one-partitioned dataset of the combined four plastid markers (thus excluding nrITS) (Table 2, Online Resource 2). The best-fitting substitution models were selected according to the AIC criterion in jModeltest. Selected substitution model for the nuclear ribosomal data was GTR + Γ and for the combined plastid dataset a symmetrical model with gamma distribution (SYM + Γ). The reconstruction of phylogeny based on datasets 3 and 4 was conducted using maximum likelihood analyses with 1000 rapid bootstrap replicates in RAxML. Since the SYM + Γ model is not available in RAxML, the more complex substitution model GTR + Γ was set for the combined plastid dataset. In addition, datasets 3 and 4 were analyzed using Bayesian inference of phylogeny and substitution models GTR + Γ for the nrITS partition and SYM + Γ for the combined plastid markers. MCMC runs were set to six million generations, and generated trees were sampled every 1000 generations. AWTY and Tracer were applied to confirm the convergence of the two MCMC runs. Phylogenies were generated by summarizing the sampled trees in a majority rule consensus tree, and the initial 600 trees were deleted as burn-in. Supported topologies from the maximum likelihood and the Bayesian inference analyses of datasets 3 and 4 were compared to assess incongruence between the nrITS marker and the combined plastid markers.
Estimation of divergence times
Divergence times of clades in the Chinese lianoid Gnetum clade were estimated based on the two-partitioned dataset of five markers (dataset 3, Table 2) in a Bayesian framework using BEAST version 1.8 (Drummond et al. 2012) via CIPRES. Since the SYM + Γ model is not available in BEAST, the more complex substitution model GTR + Γ was set for both partitions. Four alternative settings were tested: (a) a strict clock model with a pure birth process as tree prior (Yule 1925; Gernhard 2008); (b) a strict clock model with a birth–death speciation process as the tree prior (Kendall 1948; Stadler 2009); (c) an uncorrelated lognormal clock model with a pure birth process; and (d) an uncorrelated lognormal clock with a birth–death speciation process. The optimal setting was selected using a posterior simulation-based analog of the Akaike information criterion (AICM, Raftery et al. 2006; Baele et al. 2012) implemented in Tracer. Two independent MCMC runs were performed and run for 60 million generations with the sampling frequency of 20,000. A random tree was selected as the starting tree in all analyses. Nodes were calibrated to absolute time using normally distributed age priors, with monophyly enforced, based on results in Hou et al. (2015; Table 3). Separate runs excluding the data were also run, and the resulting effective prior distributions were assessed and compared to those specified. Effective sampling sizes for each Bayesian parameter were assessed in Tracer. In each run, the initial 300 trees were discarded as burn-in. The remaining sampled trees from two independent runs were combined in LogCombiner version 1.8 included in the BEAST package. The ultrametric tree was summarized with median node heights and maximum clade credibility from the sampled trees using TreeAnnotator version 1.8 included in the BEAST package.
Morphological studies
Morphological studies of the Chinese lianoid species were performed using the 11 currently accepted names of species occurring in China and adjacent regions as a framework (Table 3). Forty-one herbarium sheets of type material were studied (of G. cleistostachyum, G. gracilipes, G. hainanense, G. luofuense and G. pendulum in PE and of G. catasphaericum and G. giganteum in PEM) (herbarium acronyms following Thiers 2016). Type material (40 specimens) of the remaining species was reviewed through Global Plants on JSTOR (http://plants.jstor.org/) as well as from requested images from the type localities. In addition, 126 herbarium sheets of non-type material were studied during visits in several additional herbaria in China and elsewhere (A, B, E, HK, HITBC, IBK, KUN, P, PE, PEM, NAS and S). Species determinations of non-type material were verified using available information (protologues, keys and comparison with type material). The gross morphology of specimens was studied by eye and using a dissecting microscope. Most species were also studied in their natural environments in the field. The type localities of G. giganteum and G. catasphaericum in Guangxi were visited. Localities near the type localities of G. gracilipes, G. hainanense and G. luofuense were also visited. Twenty-one characters were defined, seven from vegetative plant parts (characters 1–7), six from male reproductive parts (characters 8–13) and eight from female reproductive parts (characters 14–21). The state of each character was noted for each specimen, and data were summarized for each of the 11 currently accepted names (Table 3). Based on the results, species boundaries were subsequently assessed and taxonomic revisions were made accordingly.
Results
Plastid genome assembly and sequence variation detection
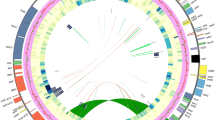
The eleven specimens of Gnetum, for which the entire plastid genome was sequenced, generated between 13,547,250 and 77,910,472 paired-end reads with the average read length of 126 bp. A total of 190,899 to 2,003,005 paired-end reads were mapped onto the reference plastid genome, resulting in the average coverage number of 298–2156. Numbers of contigs assembled de novo varied from four to nine with the maximum length 29,004 to 65,957 bp. After manual modification, 11 plastid genomes of Gnetum were completed, with the length between 114,238 bp (G. gnemon) and 114,983 bp (G. parvifolium) (Online Resource 1). Plastid genomes of Chinese lianoid Gnetum consist of 115 genes (Online Resource 3), of which 66 were protein-coding genes, eight were ribosomal RNA (rRNA) genes and 40 were transfer RNA (tRNA) genes. In addition, sequence variations across the five Gnetum plastid genomes were revealed in the identity map (Fig. 2). A total number of 9345 variable sites were detected, consisting of 5600 polymorphisms (60 %) and 3745 insertions and/or deletions (40 %). Protein-coding regions revealed a conserved pattern in the alignment with the notable exception of matK, ycf1 and ycf2 where sequence variations existed. Non-protein-coding regions, consisting of intergeneric regions, introns, rRNA and tRNA, exhibited higher variability in the alignment, especially the intergeneric regions psbD-trnT, trnT-trnK, trnN-ycf1, ycf1-trnP, trnH-trnI, rpl36-rps11, psaC-trnN, psbB-rps12 and trnF-trnV.
Identity plots of plastid genome comparison of five species of Gnetum plotted against the reference genome of G. parvifolium (GenBank ID, NC_011942) in mVISTA. Pairwise comparisons and sequence identity are visualized on the y-axis from 50 to 100 % identity across the plastid genome. All genes are annotated and shown with directions (gray arrows), protein-coding regions (purple), tRNA and rRNA regions (light blue), intron and intergeneric regions (pink) and Sanger sequenced regions (green)
Phylogenetic analyses of NGS data
An alignment comprising 66 protein-coding genes (dataset 2; 64,150 aligned sites) was extracted from the full alignment of entire plastid genomes (dataset 1; 116,201 aligned sites). Phylogenetic analyses of the two datasets revealed highly congruent results, with regard to topology and statistical support, in both the maximum likelihood analysis and the Bayesian analysis. Detailed results from the analyses of dataset 1 are reported in Fig. 3. Bootstrap values higher than 70 and posterior probabilities larger than 0.95 are considered supported (Alfaro et al. 2003). Specimens that represent G. parvifolium comprise a clade [clade 2, BS (bootstrap support) = 100, PP (posterior probability) = 1.0]. Within G. parvifolium, specimen C50 is sister to the remaining two specimens (clade 3, BS = 100, PP = 1.0). Gnetum parvifolium is sister to all the remaining Chinese lianoid specimens (clade 5, BS = 100, PP = 1.0). Clade 5 includes two specimens of G. hainanense (clade 4, BS = 100, PP = 1.0) sister to a group including specimens of G. montanum and G. pendulum (clade 7, BS = 100, PP = 1.0). Gnetum montanum (clade 6, BS = 100, PP = 1.0) is sister to G. pendulum (clade 8, BS = 98, PP = 1.0).
Phylogenetic relationships of 11 Gnetum specimens based on maximum likelihood (ML) and Bayesian inference (BI) analyses of the entire plastid genomes (dataset 1). The tree was rooted using two specimens of G. gnemon as outgroup. Numbers shown represent support values obtained from 1000 rapid bootstrap replicates from the ML analyses (left) and Bayesian posterior probabilities (right)
Phylogenetic analyses of Sanger sequencing data
The 66-taxon dataset of Sanger sequencing data comprised a total of 4690 aligned sites (dataset 3), including 3675 aligned sites for the four plastid markers (dataset 4), and 1015 aligned sites for the nrITS. Detailed results from each analysis are reported in Fig. 4a, c (dataset 3) and Fig. 4b, d (dataset 4). There were no statistically supported conflicts between results from an analysis of the nrITS alone (not shown), and the concatenation of four plastid markers used in our study. Maximum likelihood and Bayesian inference analyses of datasets 3 and 4 revealed generally congruent results with regard to both topology and statistical support. These results are also highly congruent with those obtained from the analysis of NGS data (Fig. 3) and from results of the dating analysis (Fig. 5). Noteworthy, however, is that the clade comprising four G. montanum specimens yielded slightly lower statistic support when the nrITS marker was included in the Bayesian inference analyses (PP = 0.99 vs. 0.92, Fig. 4). The results considered below are those from the analyses of dataset 3 (including both nrITS and plastid markers).
Phylogenetic relationships of Gnetum. a Results based on a two-partitioned dataset (dataset 3), including one nuclear ribosomal marker and the combined four plastid markers. b Results based on a one-partitioned dataset (dataset 4), including the combined four plastid markers. Analyses are conducted using maximum likelihood and Bayesian inference methods, and with G. africanum as outgroup. c–d Corresponding phylograms are presented. Values on nodes represent statistical support obtained from 1000 rapid bootstrap replicates from the ML analyses (left), and Bayesian posterior probabilities (right). Thick branches indicate bootstrap support ≥95 % and/or posterior probabilities ≥0.95

modified from Hansen et al. (2013)
Chronogram resulting from the divergence time analysis using BEAST version 1.8 (Drummond et al. 2012) of a two-partitioned dataset (dataset 3), including the nuclear ribosomal marker and the combined four plastid markers. Bars indicate maximum clade credibility intervals, estimated by the 95 % highest posterior density (HPD), for each node. Nodes labeled with yellow circles are nodes on which prior age estimates are applied based on results in Hou et al. (2015) (see also Table 4). Median divergence ages of Chinese lianoid Gnetum are shown with 95 % HPD in brackets. A temperature curve from the Miocene to the present is given,
Ten specimens that represent G. parvifolium form a clade (clade P, BS = 100, PP = 1.0). Relationships within the clade are resolved but generally poorly supported. Gnetum parvifolium is sister to the remaining taxa of the Chinese lianoid clade (clade R, BS = 100, PP = 1.0). Clade R comprises two specimens of G. formosum, which are excluded from clade S (BS = 100, PP = 1.0) comprising all remaining specimens. Clade S exhibits a trichotomy that consists of the clades V (BS = 99, PP = 1.0), W (BS = 97, PP = 1.0) and T (BS = 98, PP = 1.0). Clade V includes five specimens representing G. catasphaericum. Clade W includes seven specimens of G. pendulum, four specimens of G. montanum, three specimens of G. giganteum and two specimens of G. gracilipes. The clade comprising the four specimens of G. montanum was well supported in the Bayesian inference phylogeny (PP = 1.0) but not in the maximum likelihood analyses. Remaining relationships in clade W were either unresolved or poorly supported. Clade T comprises five specimens of G. luofuense and twelve specimens of G. hainanense. As with clade W, resolution in this clade is poor and the two taxon groups do not constitute separate clades.
Divergence time estimates
The birth–death prior combined with an uncorrelated (lognormal) relaxed clock (scenario d) had the best fit to the data as estimated from bootstrap simulation in AICM, and these settings were used in all subsequent divergence time analyses. Effective age priors were consistent with those specified. Divergence time estimates are shown in Fig. 5 in the form of a chronogram calibrated against the geological time scale (Gradstein et al. 2012), and in Table 4 where mean divergence and credibility intervals, as estimated by the 95 % HPD intervals, are given for major clades. The mean crown age of Chinese lianoid Gnetum was estimated to 21 Ma (clade O, 95 % highest posterior density [HPD]: 11–34 Ma corresponding to the Oligocene to early Miocene). The mean crown age of clade P (i.e., G. parvifolium) was 6 Ma (HPD: 2–11 corresponding to the late Miocene to Pliocene), which is younger than the mean crown age of clade R dated at 9 Ma (HPD: 4–17 early Miocene to Pliocene). Following a divergent event, clade Q (i.e., G. formosum) emerged at 2 Ma (HPD: 0–8, late Miocene to extant) and clade S (i.e., the remaining lianoid species) at 6 Ma (HPD: 3–10, late Miocene to Pliocene). The mean age of clade V (i.e., G. catasphaericum) was estimated at 4 Ma (HPD: 2–7 Ma; latest Miocene to Pliocene), which is older than the mean crown ages of clade T (i.e., G. luofuense) at 3 Ma (HPD: 1–5 Ma; Pliocene to Pleistocene) and clade W (i.e., G. pendulum and G. montanum) at 2 Ma (HPD: 1–4 Ma, Pliocene to Pleistocene).
Morphological studies and taxonomic conclusions
Based on the comparative studies of gross morphology, together with the results from our molecular analyses, the Chinese lianoid clade is proposed to comprise six species, i.e., G. catasphaericum, G. formosum, G. luofuense, G. montanum, G. parvifolium and G. pendulum. Gnetum hainanense C.Y.Cheng ex L.K.Fu, Y.F.Yu & M.G.Gilbert is included in and synonymized with G. luofuense C.Y.Cheng. Gnetum giganteum H.Shao and G. gracilipes C.Y.Cheng are included in and synonymized with G. pendulum C.Y.Cheng. New species circumscriptions and studied material are listed in Online Resource 4. Keys to the six recognized Chinese species (modified from Fu et al. 1999a), one based on male plants and one on female plants, are presented in the section of taxonomic treatment.
Discussion
According to the currently accepted classification, there are 11 lianoid species in China. However, based on our phylogenetic results and morphological studies, we conclude that there are only six lianoid species in the Chinese Gnetum clade (Fig. 5; Online Resource 4): G. parvifolium, G. catasphaericum, G. formosum, G. luofuense, G. montanum and G. pendulum. Remaining names are synonymous with other taxa. Distinct structural differences of plastid genomes were found among Chinese species of Gnetum. Interestingly, these differences are not only evident between the arborescent and the lianoid species, but also within the clade of Chinese lianoid species. Species delimitations are well resolved using the entire plastid genomes, but for more economic extraction of data from many specimens we developed four new plastid markers. Our morphological investigations corroborate the results from molecular data (with the possible exception of G. catasphaericum) and show for example that species with substantial intraspecific genetic divergence, like G. parvifolium, also have distinct intraspecific morphological variation that appears correlated with geographic regions. Similarly, previously recognized species that cannot be separated based on our molecular data (e.g., G. pendulum, G. giganteum and G. gracilipes), also cannot be separated morphologically.
Estimated mean crown ages of species fall within the Pliocene to Pleistocene epochs. Only G. parvifolium may be slightly older (latest Miocene). The entire clade of Chinese lianoid Gnetum is from the Oligocene to Miocene and deep divergence events in the clade took place in the Miocene (Fig. 5). In the Miocene, tropical and subtropical areas expanded in South China, probably due to elevated mean temperature and annual precipitation (Yao et al. 2011), with expansion of broad-leaved forests as a consequence (Zhao et al. 2004). These transitions should have facilitated speciation and geographic expansion in the Chinese lianoid clade of Gnetum and may explain the largely overlapping geographic distribution of extant species. From the Pliocene, the climate gradually became cooler and drier (Ravelo et al. 2004; Xu et al. 2004; Kou et al. 2006) (Fig. 5b), and based on the long stem lineages to several of the extant species, it seems possible that extinction affected the clade during the late Miocene and Pliocene.
Phylogenetic relationships and species delimitations
Clade P
Our phylogenetic analyses of both NGS data and Sanger sequence data strongly support that included specimens of G. parvifolium are monophyletic and that G. parvifolium is sister to remaining species of the lianoid Chinese clade (Figs. 3, 4, 5). The analyses also indicate considerable sequence divergence among the 10 included specimens of G. parvifolium (Fig. 4c, d), although there is only weak or no support for the relationships between the different specimens. The pattern seems to corroborate results in an earlier study where G. parvifolium was shown to possess high levels of genetic intraspecific diversity (Huang et al. 2010). Our morphological studies, based on material collected across the entire geographic distribution of the species (Fujian, Guangdong, Guangxi, Hainan, Hunan, Jiangxi and Hong Kong), further support the independent status of G. parvifolium. A combination of characters separates it from remaining Chinese lianoid Gnetum (Table 3), i.e., small leaf size and inconspicuous venation, short petioles (Fig. 1d) and sessile, ellipsoid seeds (Fig. 1g, i). Leaf and seed sizes of the specimens collected in Guangxi are, however, considerably larger than seen in specimens collected in Fujian, Guangdong and Hong Kong, indicating that intraspecific variation also of gross morphology occurs in G. parvifolium. This is further supported by variation in their average pollen size as reported by Yao et al. (2004). They documented G. parvifolium to have smaller average pollen size than G. cleistostachyum, G. hainanense, G. luofuense, G. montanum and G. pendulum (Yao et al. 2004).
Clade Q
Gnetum formosum was described by Markgraf (1930) on material from Vietnam, but the recognition of G. formosum and its separation from G. parvifolium has, at least in China, been unclear in previous taxonomic work. Markgraf (1930) considered G. formosum to be endemic to Vietnam. Also, Fu et al. (1999a) viewed G. formosum to be known with certainty only from Vietnam and did not recognize it as a Chinese species in their treatment of the Flora of China (Fu et al. 1999a). Female material, collected in Hainan (China) under the name G. formosum, was not accepted by Fu et al. (1999a) who considered the material not to differ significantly from plants of G. parvifolium. Wu and Chen (1978), in their Flora of China treatment, did not recognize G. formosum as a Chinese species either, and they even raised doubts about the Vietnamese material, questioning if not also this should be included under G. parvifolium (Wu and Chen 1978). In contrast, results from our molecular analyses reveal strong support for a separation of G. formosum and G. parvifolium, and the included specimens of G. formosum are resolved sister to the other species in clade R rather than falling with G. parvifolium (Figs. 4, 5). This is true for the Chinese specimen from Hainan (G. formosum 14), as well as for a specimen from Laos (G. formosum 21). These results not only firmly establish the separation of G. formosum from G. parvifolium, but in addition expand the distribution of G. formosum to also include southern China (Hainan) as well as Laos (in the Vietnamese peninsula). Our investigation of gross morphology lend further support for these results; specimens of G. formosum differ from G. parvifolium by having a black or dark brown stem surface, larger leaves, longer petioles and narrowly oblong or fusiform seeds (Fig. 1j). They can also be distinguished from the other lianoid species by having 6–10 involucre collars placed in female spikes and sessile fusiform seeds (Table 3). Of potential concern is the lack of support for monophyly of G. formosum in our non-clock model-based molecular analyses (Fig. 4a, b). However, the two specimens are resolved together with moderate support in the relaxed clock model-based analysis (PP = 0.92; Fig. 5), they are with good support separated both from G. parvifolium and from remaining species in clade S in all analyses (Figs. 4, 5), and they are clearly distinguishable based on morphology (Table 3). We therefore consider it fully justified to treat G. formosum as an independent species. Our result further indicates that its presence in both Laos and China (Hainan) has previously been overlooked.
Clade V
Gnetum catasphaericum was originally described by Shao (1994), and the type material was collected in Shangsi, Guangxi (China). However, the species status of G. catasphaericum has previously never been carefully assessed, probably due to difficulties of accessing plant material. Fu et al. (1999a), for example, indicated that they had not seen any material of this species. For the purpose of the present study, we collected new material of G. catasphaericum from three different localities in Guangxi (China), including the type locality. We also reassessed the identity of two previously collected herbarium specimens, one collected in Yunnan (China), and one collected in Vietnam, and the results of our phylogenetic analyses clearly show that all five specimens form a strongly supported monophyletic group (Fig. 4a, b). The analyses also indicate considerable sequence divergence among the investigated specimens (Fig. 4c, d). Unlike the situation in G. parvifolium, there is good support for the relationships among different specimens, and the relationships reflect geographical patterns. The three specimens from Guangxi (specimens 32, 33 and 44) are grouped together with the specimen from Yunnan (specimen 31) as sister. Sister to all the Chinese specimens is the specimen from Vietnam (specimen 24). Whether or not these patterns indicate the presence of cryptic and as yet undescribed species in this group should be investigated further, but this will require a more extensive sample of G. catasphaericum than included in the present analyses. Morphologically, G. catasphaericum can be distinguished from the other Chinese lianoid species by possessing relatively few female units on each collar (5–8) (Fig. 1f), fewer basal hairs and oblong, subglobose seeds with contracted seed stipes 2–6 mm long when dried (Fig. 1k; Table 3). In addition, intraspecific variation of reproductive characters is detected among the four specimens of G. catasphaericum used for our molecular studies. For example, the number of fertile ovules in specimen 32 is 5–6, which is fewer than 7–8 of specimens 44 collected in another site of Guangxi.
Clade W
The well-supported clade W comprises representatives of four previously recognized species, G. pendulum, G. giganteum, G. gracilipes and G. montanum. Our molecular analyses included four Chinese specimens of G. montanum, two from Guangxi (specimens 105, 106) and one from Yunnan (specimen 17), as well as a specimen from Thailand (specimen 29). They are always resolved as monophyletic, although only some analyses (Figs. 4b, 5) provide strong support for the G. montanum clade. Gnetum montanum was described by Markgraf (1930), and he listed no less than 43 different syntypes in the original protologue. The listed types indicate a broad geographical distribution of the species and include material collected in China (Guangxi, Yunnan), India (Assam, Sikkim), Burma, Thailand and Vietnam. However, Fu et al. (1999a) pointed at several problems regarding the application of the name G. montanum. For example, the 43 syntypes are morphologically highly variable and include specimens not only of this taxon but also of G. latifolium Blume (Fu et al. 1999a). They also pointed out that the name G. montanum commonly has been used in SE China, especially in Hong Kong, for material of G. luofuense. Following the protologue as well as current usage of the name, Fu et al. (1999a) considered G. montanum as best characterized by having male involucral collars with fewer microsporangia and smaller stipitate seeds. They applied the name G. montanum accordingly but at the same time argued that this (current) application of the name should be conserved by selecting an appropriate lectotype (Fu et al. 1999a). This, however, they never did. Morphologically, the species is distinct as assessed by our observations, with more collars on the male spikes (Fig. 1c), fewer reproductive units on female spikes and relatively small stipitate seeds (Fig. 1l) compared to other Chinese species. Based on this, and the strong support for monophyly of the species in at least some of our analyses of molecular data, we find it justified to recognize G. montanum. We also agree with Fu et al. (1999a) that lectotypification is warranted. Unfortunately, we have not been able to see any of the type specimens from China listed by Markgraf (1930), because they are located in herbaria outside of China not visited by us. However, the specimen from Thailand included in our molecular analyses was collected from the exact same area (Chieng Mai) as was Kerr n. 602, a syntype specimen listed by Markgraf (1930). We have therefore chosen Kerr 602 (K) as the designated lectotype for G. montanum (see nomenclature below), conserving the current application of the name as suggested by Fu et al. (1999a).
The remaining specimens of clade W represent three additional species that have been recognized in treatments of the Chinese flora: G. pendulum, G. gracilipes and G. giganteum (Cheng et al. 1975; Wu and Chen 1978; Fu et al. 1999a). The first two, G. pendulum and G. gracilipes, were originally described by Cheng et al. (1975) on collections from Yunnan (G. pendulum) and from Guangxi (G. gracilipes). However, the validity of the original diagnoses was questioned by Fu et al. (1999b) as Cheng et al. (1975) include no direct information on the species being described. They only indicate how some other species differ from it, and both were compared to species found well outside of China. Gnetum pendulum was compared to G. oblongum Markgr., a species described by Markgraf (1930) from the Chittagong Hill Tracts in Bangladesh, and G. gracilipes was compared to G. arboreum Foxw., a species that does not even have a lianoid habit and was described from the Philippines by Foxworthy (1911). Both comparisons are of questionable value, but G. pendulum and G. gracilipes have remained recognized in more recent work (Wu and Chen 1978; Fu et al. 1999a). One feature that G. pendulum (Fig. 1m) and G. gracilipes (Fig. 1n) seem to share is long-stipitate seeds, although Cheng et al. (1975) considered the character so variable in G. pendulum that they described different forms: G. pendulum f. intermedium C.Y. Cheng and G. pendulum f. subsessile C.Y. Cheng. The first name, G. pendulum f. intermedium is invalid as two different types (male and female) were designated in the original protologue (Cheng et al. 1975).
Fu et al. (1999a) considered G. pendulum as closely related to, and G. gracilipes as showing many similarities to, G. latifolium. This is a putatively widespread and highly variable species described by Blume (1834) from Java, Indonesia, for which a large number of different forms and varieties have been described. Our analyses clearly indicate a different pattern (Fig. 4a, b). The included specimen of G. latifolium is resolved together with other species from Southeast Asia in clade K, nowhere close to the G. pendulum and G. gracilipes specimens. Also in previous work, including additional specimens of G. latifolium (Won and Renner 2006; Hou et al. 2015) are specimens of G. latifolium monophyletic and included in the equivalent of our clade K, i.e., clearly separated from the Chinese lianoid clade.
The third species included in clade W is G. giganteum, described by Shao (1994) based on type material collected in Guangxi. It was described to differ from other species by having large, sessile and broadly ellipsoid seeds, and female involucral collars separated by up to 3 cm when mature (Shao 1994). The status also of this species has not previously been carefully assessed. There are very few collections of G. giganteum in Chinese herbaria, and Fu et al. (1999a), for example, indicated that they had not seen any material of this species.
Based on our morphological work, it is clear that the three taxonomic entities G. pendulum, G. gracilipes and G. giganteum share characters with overlapping states and are morphologically indistinguishable (Table 3). Further, although the representing specimens are unresolved within clade W in our non-clock model-based molecular analyses (Fig. 4a, b), they form a clade in the relaxed clock model-based analysis (Fig. 5). We therefore conclude that it is relevant to assign material previously referred to any of the three names G. pendulum, G. gracilipes and G. giganteum to the same single species (i.e., G. pendulum). From a sequence divergence point of view, there is a striking pattern with almost no variation at all in the entire clade W (Fig. 4c, d). Strictly speaking, no conclusions can be drawn based on this almost complete absence of molecular variability, but a comparison with the divergence patterns seen in the G. parvifolium and G. catasphaericum clades indicates that other processes are active in clade W (and clade T) than in clades P and V regarding the exchange of genetic material among individuals.
Clade T
Also, another clade with almost no sequence divergence among included specimens even though the specimens represent more than one taxon, is identified in our analyses. The taxa concerned are G. hainanese and G. luofuense and specimens representing these two taxa are all resolved in the well-supported clade T (Figs. 4, 5). Morphologically, G. hainanese and G. luofuense can be distinguished from the other Chinese lianoid species by possessing longer male spikes (Fig. 1d), more fertile units on each collar (Fig. 1e), seed ornamentation (Fig. 1h) and short-stipitate seeds (Fig. 1o, p) (Table 3). Both species were originally described by Cheng et al. (1975) based on material from China, G. hainanense from Hainan and G. luofuense from Guangdong. Their description of G. hainanense pointed, however, at both a female and a male type specimens and the name was for this reason invalid. Fu et al. (1999b) noticed this problem and supplied a valid description of G. hainanense based on the female type specimen from Cheng et al. (1975). They described G. hainanense as possessing apparently larger seeds with smooth outer seed coats (Fig. 1o) compared with smaller wrinkled seeds of G. luofuense (Fig. 1p) (Fu et al. 1999b). However, a separation of G. hainanense and G. luofuense is not supported in the analyses conducted here. Our molecular analyses indicate almost no sequence divergence whatsoever between the included specimens (Fig. 4c, d), and also the results of our morphological studies reveal no differentiation in the features investigated (Table 3). These include the number of lateral veins on the leaves, and the size and shape of the seeds, features that were used in the original protologue of G. hainanense to separate it from G. luofuense (Fu et al. 1999b). Yao et al. (2004) investigated the exine ultrastructure and size of pollen grains from Chinese species of Gnetum, and they too found no differentiation between G. hainanense and G. luofuense. We therefore consider G. hainanense synonymous to the older name G. luofuense.
Gnetum indicum (Lour.) Merr
The type material of G. indicum was collected in Vietnam in the early 1770s. The name is possibly synonymous with G. parvifolium, but the taxonomic history of G. indicum is complex and the name has been applied in several ways in the past, often in the sense of G. montanum according to Fu et al. (1999a). Markgraf (1930) argues that the type material is vegetative and difficult to interpret, and we largely agree although there actually is a young seed on the sheet. The seed is cracked and important information on its original shape is not preserved. Therefore, and in the absence of DNA data from the type, we find it difficult to assess the affinity of the type material of G. indicum with certainty. Based on available information, it may represent G. parvifolium, G. formosum or a species of its own.
Gnetum cleistostachyum C.Y.Cheng
This name is invalid because both a male and a female specimen were indicated as the type specimen, and the protologue only states a few sentences on how the material differs from G. pendulum (Cheng et al. 1975). More importantly, however, very little material of this species has ever been collected and Fu et al. (1999b) considered it wise to postpone a validation of the name. Based on our morphological investigations, it seems possible that the type material of G. cleistostachyum in fact represents G. montanum. The “closed” nature of the reproductive spikes of G. cleistostachyum is most probably unimportant and a developmental difference; all known spikes of G. cleistostachyum are at an early developmental stage. There are, however, one apparent difference between G. cleistostachyum and G. montanum (the number of male reproductive units on each collar), but in the absence of additional morphological data and/or DNA data from G. cleistostachyum, we refrain from assessing whether or not the material represents a unique species.
Taxonomic notes
Characters of the seed, such as size, shape, surface ornamentation and length of the stipe, have previously been considered useful in identifying species and forma of Chinese lianoid Gnetum (Cheng et al. 1975; Wu and Chen 1978; Fu et al. 1999a). Some seed characters are, however, variable within species and/or overlap between species. For example, the fleshy seed coat may be both coarsely reticulate-wrinkled and smooth in G. luofuense; the basal part of seeds contract into a “false” stipe when dried in G. luofuense and G. catasphaericum, which then becomes indistinguishable from the “true” seed stipe of G. montanum formed during the development; and seed ornamentation of silver scales are often present in both G. luofuense and G. pendulum. Further, we find details of hairs that surround female reproductive spikes uninformative and not useful for species identification in the Chinese lianoid clade; hairs are usually brown, short and inconspicuous in these species. The new key to female specimens presented here (see below) therefore makes use of other reproductive characters (e.g., spike length, length between involucre collars and number of fertile units on each collar, see Table 3). The key to male specimens is based on characters such as spike length, number of sterile units on each collar and amount of basal hairs. Vegetative characters such as measurements of leaves size, leaf surface pattern, color of leaves when dried, are broadly applied for distinguishing species among South American species (Markgraf 1965) and Malaysian species (Markgraf 1951), but are not useful to identify Chinese lianoid Gnetum because the characters are largely overlapping (see Table 3). Nevertheless, such characters can be useful in combination with reproductive characters.
Taxonomic treatment
Gnetum parvifolium (Warb.) W.C.Cheng, Acta Phytotax. Sin. 9: 386. 1964. ≡ Gnetum scandens Roxb. var. parvifolium Warb., Monsunia 1: 196. 1900. ≡ Gnetum montanum Markgr. f. parvifolium (Warb.) Markgr., Bull. Jard. Bot. Buitenzorg Ser. III, 10(4): 468. 1930. ≡ Thoa parvifolia (Warb.) Doweld, Turczaninowia 3(4): 32. 2000. —TYPE: China, Futschan, Dec 1887, Warburg 5953 (lectotype designated here, or perhaps holotype: B barcode B100001504 [photo!]).
Gnetum formosum Markgr., Bull. Mus. Natl. Hist. Nat. Ser. II, 2: 686. 1930. —TYPE: Vietnam, La Han, Thanh hoa, 20 Aug 1920, Poilane 1689 (lectotype designated here, or perhaps holotype: P barcode P00722133 [photo!]; isolectotypes: P barcode P00722134 [photo!], P barcode P00722135 [photo!], B barcode B100001469 [photo!]).
Gnetum catasphaericum H.Shao, Guihaia 14: 297. 1994. ≡ Thoa catasphaerica (H.Shao) Doweld, Turczaninowia 3(4): 32. 2000. —TYPE: China, Guangxi, Shangsi, 8 Sep 1987, Shao 87053 (holotype: PEM [n.v.]).Footnote 1
Gnetum montanum Markgr., Bull. Jard. Bot. Buitenzorg Ser. III, 10(4): 466. 1930. ≡ Thoa montana (Markgr.) Doweld, Turczaninowia 3(4): 32. 2000. —TYPE: Thailand, Chieng Mai, Doi Sootep, 21 Apr 1909, Kerr A.F.G. 602 (lectotype designated here: K barcode K000454548; isolectotype: P barcode P00722156 [photo!]).
Gnetum pendulum C.Y.Cheng, Acta Phytotax. Sin. 13(4): 88. 1975. —TYPE: China, Yunnan, Lincang, 28 Sep 1938, Yu 17755 (lectotype designated here, or perhaps holotype: PE barcode PE00002924!).
= Gnetum pendulum C.Y.Cheng f. subsessile C.Y.Cheng, Acta Phytotax. Sin. 13(4): 88. 1975. —TYPE: China, Yunnan, Tengchong, Yi 60-1902 (lectotype designated here, or perhaps holotype: PE barcode PE00002920 [image!]).
= Gnetum giganteum H. Shao, Guihaia 14: 298. 1994. —TYPE: China, Guangxi, Shangsi, 8 Sep 1987, Shao 87058 (holotype: PEM [n.v.]) (see footnote 1).
= Gnetum gracilipes C.Y.Cheng Acta Phytotax. Sin. 13(4): 88. 1975. —TYPE: China, Guangxi, Sup Man Ta Shan, Liang 69874 (lectotype designated here, or perhaps holotype: PE barcode PE00002919!; isolectotypes: P barcode P00722134 [photo!], P barcode P00722135 [photo!], B barcode B100001469 [photo!], IBK barcode IBK00041278 [photo!]).
Gnetum luofuense C.Y.Cheng, Acta Phytotax. Sin. 13(4): 89. 1975. —TYPE: China, Guangdong, Luofushan, 10 Aug 1930, Chen 41527 (lectotype designated here, or perhaps holotype: PE 00002913!; isolectotype: MO barcode MO954421 [photo!]).
= Gnetum hainanense C.Y.Cheng ex L.K.Fu, Y.F.Yu & M.G.Gilbert, Novon 9(2): 187, 1999 ≡ Thoa hainanensis (C.Y.Cheng ex L.K.Fu, Y.F.Yu & M.G.Gilbert) Doweld, Turczaninowia 3(4): 33. 2000. —TYPE: China, Hainan Island, Hung Mo Shan, 22 Jul 1929, Tsang et Fung 554 (holotype: PE barcode PE00002917!; isotypes: A barcode A00022589 [photo!], K barcode K000454089 [photo!], PE barcode PE00002918).
Key based on male plants
1a. Shrubs or small trees; leaves yellowish when dried; sterile ovules and axis visible between involucral collars … G. gnemon
1b. Liana; leaves dark brown or black when dried; spikes condensed with axis invisible between involucral collars … 2
2a. Sterile ovules 7–12; surrounding basal hairs few … 3
2b. Sterile ovules 13–22; surrounding basal hairs dense … 4
3a. Male spike 0.5–1.5 cm in length; inconspicuous lateral veins on the adaxial surface of leaves … G. parvifolium
3b. Male spike ca. 2 cm in length; apparent lateral veins on the adaxial surface of leaves … G. catasphaericum
4a. Male spike 1–1.5 cm in length; male spikes branching three to five times … G. pendulum
4b. Male spike 2–3.5 cm in length; male spike simple, or once or twice branching … 5
5a. Involucral collars 12–15; male reproductive units 60–80 … G. luofuense
5b. Involucral collars 16–25; male reproductive units 20–45 … G. montanum
Key based on female plants
1a. Shrubs or small trees; leaves yellowish when dried; involucral collars flat … G. gnemon
1b. Liana; leaves dark brown or black when dried; involucral collars funnel-shaped … 2
2a. Seed sessile; inconspicuous lateral veins on the adaxial surface of leaves … 3
2b. Sterile ovules 13–22; surrounding basal hairs dense … 4
3a. Seed 1.3–1.8 cm in length, 0.4–1 cm in width; leaf 4–11 cm in length, 2–4 cm in width … G. parvifolium
3b. Seed 2–2.5 cm in length, 0.8–0.9 cm in width; leaf 2–2.5 cm in length, 0.5–0.9 cm in width … G. formosum
4a. Fertile ovules 9–13; collars 11–25 mm apart … 5
4b. Fertile ovules 5–8; collars 6–10 mm apart … 6
5a. Seeds 3–4 cm in length, 1.7–1.8 cm in width; seed stipe 8–30 mm long … G. pendulum
5b. Seeds 1.8–2.5 cm in length, 1.3–1.7 cm in width; seed stipe contracted into 2–5 mm long when mature … G. luofuense
6a. Seeds oblong, subglobose; seed stipe contracted into 2–6 mm long when mature … G. catasphaericum
6b. Seeds oblong, subglobose; seed stipe contracted into 2–6 mm long when mature … G. montanum
Notes
The type sheets assigned in the protologue were not found while visiting the PEM herbarium.
References
Alfaro ME, Zoller S, Lutzoni F (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molec Biol Evol 20:255–266. doi:10.1093/molbev/msg028
Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV (2012) Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molec Biol Evol 29:2157–2167. doi:10.1093/molbev/mss084
Blume CL (1834) De novis quibusdam plantarum familiis expositio et olim jam expositarum enumeratio. Tijdschr Natuurl Gesch Physiol 1:131–162
Cheng WC (1964) A new name for a species of Gnetum L. Acta Phytotax Sin 9:386
Cheng WC, Fu LK, Cheng CY (1975) Gymnospermae sinicae [In Chinese]. Acta Phytotax Sin 13:88–89
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Meth 9:772. doi:10.1038/nmeth.2109
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510. doi:10.1038/nrg3012
Deschamps S, Campbell MA (2010) Utilization of next-generation sequencing platforms in plant genomics and genetic variant discovery. Molec Breed 25:553–570. doi:10.1007/s11032-009-9357-9
Doyle J (1991) DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer, Berlin, pp 283–293
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molec Biol Evol 29:1969–1973. doi:10.1093/molbev/mss075
Ekblom R, Galindo J (2011) Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107:1–15. doi:10.1038/hdy.2010.152
Foxworthy FW (1911) Philippine gymnosperms. Philipp J Sci C 6:149–178
Fu LK, Yu YF, Gilbert MG (1999a) Gnetaceae. In: Wu ZY, Raven PH (eds) Flora of China. Missouri Botanical Garden Press, St. Louis, pp 102–105
Fu LK, Yu YF, Gilbert MG (1999b) Validation of an invalidly described species of Gnetum (Gnetaceae) from Hainan Island, China. Novon 9:187–189. doi:10.2307/3391796
Gernhard T (2008) The conditioned reconstructed process. J Theor Biol 253:769–778. doi:10.1016/j.jtbi.2008.04.005
Gradstein FM, Ogg G, Schmitz MD (2012) The chronostratigraphic scale 2012. Elsevier, Boston. doi:10.1016/B978-0-444-59425-9.00004-4
Hansen J, Sato M, Russell G, Kharecha P (2013) Climate sensitivity, sea level and atmospheric carbon dioxide. Phil Trans Roy Soc A 371:20120294. doi:10.1098/rsta.2012.0294
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6:e19254. doi:10.1371/journal.pone.0019254
Hou C, Humphreys AM, Thureborn O, Rydin C (2015) New insights into the evolutionary history of Gnetum (Gnetales). Taxon 64:239–253. doi:10.12705/642.12
Huang SB, Hu YH, Wu D, Tian Q, Li HQ (2010) Genetic diversity of Gnetum parvifolium of Fujian by ISSR markers [In Chinese]. Guihaia 30:601–607
Huang H, Shi C, Liu Y, Mao S-Y, Gao L-Z (2014) Thirteen Camellia plastid genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol 14:151. doi:10.1186/1471-2148-14-151
Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104:19369–19374. doi:10.1073/pnas.0709121104
Katoh K, Misawa K, Kuma KI, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066. doi:10.1371/journal.pone.0019254
Kearse M, Moir R, Wilson A, Stones Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi:10.1093/bioinformatics/bts199
Kendall DG (1948) On the generalized” birth-and-death” process. Ann Math Stat 19:1–15
Kou XY, Ferguson DK, Xu JX, Wang YF, Li CS (2006) The reconstruction of paleovegetation and paleoclimate in the Late Pliocene of West Yunnan, China. Clim Change 77:431–448. doi:10.1007/s10584-005-9039-5
Kubitzki K (1990) Gnetaceae. In: Kramer KU, Green PS (eds) The families and genera of vascular plants, vol 1., Pteridophytes and gymnospermsSpringer, Berlin, pp 383–386. doi:10.1007/978-3-662-02604-5_71
Lin CP, Huang JP, Wu CS, Hsu CY, Chaw SM (2010) Comparative plastid genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol 2:504–517. doi:10.1093/gbe/evq036
Liu AL, Yang F, Zhu M, Zhou D, Lin M, Lee SM, Wang YT, Du GH (2010) In vitro anti-influenza viral activities of stilbenoids from the lianas of Gnetum pendulum. Pl Med 76:1874–1876. doi:10.1055/s-0030-1250030
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis, version 3.01. http://mesquiteproject.org/
Mariac C, Scarcelli N, Pouzadou J, Barnaud A, Billot C, Faye A, Kougbeadjo A, Maillol V, Martin G, Sabot F (2014) Cost-effective enrichment hybridization capture of plastid genomes at deep multiplexing levels for population genetics and phylogeography studies. Molec Ecol Resources 14:1103–1113. doi:10.1111/1755-0998.12258
Markgraf F (1930) Monographie der Gattung Gnetum Ser. 3. Bull Jard Bot Buitenzorg 10:407–511
Markgraf F (1951) Gnetaceae. In: Steenis CGGJ (ed) Flora Malesiana ser 1, vol 4. Djakarta, Noordhoff-Kolff, pp 336–347
Markgraf F (1965) New discoveries of Gnetum in tropical America. Ann Missouri Bot Gard 52:379–386. doi:10.2307/2394800
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047. doi:10.1093/bioinformatics/16.11.1046
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science Gateway for inference of large phylogenetic trees. Gateway computing environments workshop (GCE). IEEE
Moore MJ, Hassan N, Gitzendanner MA, Bruenn RA, Croley M, Vandeventer A, Horn JW, Dhingra A, Brockington SF, Latvis M (2011) Phylogenetic analysis of the plastid inverted repeat for 244 species: insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Int J Pl Sci 172:541–558. doi:10.1086/658923
Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ (2011) Plastid genome sequences from total DNA for plant identification. J Pl Biotech 9:328–333. doi:10.1111/j.1467-7652.2010.00558.x
Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24:581–583. doi:10.1093/bioinformatics/btm388
Parks M, Cronn R, Liston A (2009) Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of plastid genomes. BMC Biol 7:84. doi:10.1186/1741-7007-7-84
Powell W, Morgante M, McDevitt R, Vendramin G, Rafalski J (1995) Polymorphic simple sequence repeat regions in plastid genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA 92:7759–7763
Price RA (1996) Systematics of the Gnetales: a review of morphological and molecular evidence. Int J Pl Sci 157:S40–S49. doi:10.1086/297402
Provan J, Powell W, Hollingsworth PM (2001) Plastid microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147. doi:10.1016/S0169-5347(00)02097-8
Raftery AE, Newton MA, Satagopan JM, Krivitsky PN (2006) Estimating the integrated likelihood via posterior simulation using the harmonic mean identity. April 2006, Memorial Sloan-Kettering Cancer Center, Dept. of Epidemiology & Biostatistics Working Paper Series. Working Paper 6. http://biostats.bepress.com/mskccbiostat/paper6
Rambaut A, Drummond AJ (2003) Tracer: MCMC trace analysis tool. University of Oxford. http://tree.bio.ed.ac.uk/software/tracer/
Ravelo AC, Andreasen DH, Lyle M, Lyle AO, Wara MW (2004) Regional climate shifts caused by gradual global cooling in the Pliocene epoch. Nature 429:263–267. doi:10.1038/nature02567
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Rounsley S, Marri PR, Yu Y, He R-F, Sisneros N, Goicoechea JL, Lee SJ, Angelova A, Kudrna D, Luo M-Z (2009) De novo next generation sequencing of plant genomes. Rice 2:35–43. doi:10.1007/s12284-009-9025-z
Rydin C, Pedersen KR, Friis EM (2004) On the evolutionary history of Ephedra: cretaceous fossils and extant molecules. Proc Natl Acad Sci USA 101:16571–16576. doi:10.1073/pnas.0407588101
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res 33:W686–W689. doi:10.1093/nar/gki366
Shao H (1994) Two new species of Gnetum L. from Guangxi. Guihaia 14:297–299
Staden R (1996) The Staden sequence analysis package. Molec Biotechnol 5:233–241. doi:10.1007/BF02900361
Stadler T (2009) On incomplete sampling under birth–death models and connections to the sampling-based coalescent. J Theor Biol 261:58–66. doi:10.1016/j.jtbi.2009.07.018
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi:10.1093/bioinformatics/btu033
Straub SC, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A (2012) Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Amer J Bot 99:349–364. doi:10.3732/ajb.1100335
Thiers B (2016) Herbariorum, Index: a global directory of public herbaria and associated staff, New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/ih/
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Won H, Renner SS (2003) Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA 100:10824–18029. doi:10.1073/pnas.1833775100
Won H, Renner SS (2005a) The plastid trnT–trnF region in the seed plant lineage Gnetales. J Molec Evol 61:425–436. doi:10.1007/s00239-004-0240-3
Won H, Renner SS (2005b) The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Molec Phylogen Evol 36:581–597. doi:10.1016/j.ympev.2005.03.011
Won H, Renner SS (2006) Dating dispersal and radiation in the gymnosperm Gnetum (Gnetales)—clock calibration when outgroup relationships are uncertain. Syst Biol 55:610–622. doi:10.1080/10635150600812619
Wu ZY, Chen SC (1978) Gnetaceae. In: Cheng CY, Fu LK (eds) Flora republicae popularis sinicae, vol 7 [In Chinese]. Science Press, Beijing, pp 490–502
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. doi:10.1093/bioinformatics/bth352
Xu JX, Ferguson DK, Li CS, Wang YF, Du NQ (2004) Climatic and ecological implications of Late Pliocene palynoflora from Longling, Yunnan, China. Quatern Int 117:91–103. doi:10.1016/S1040-6182(03)00119-8
Yao CS, Lin M (2005) Bioactive stilbene dimers from Gnetum cleistostachyum. Nat Prod Res 19:443–448. doi:10.1080/14786410412331271041
Yao YF, Xi YZ, Geng BY, Li CS (2004) The exine ultrastructure of pollen grains in Gnetum (Gnetaceae) from China and its bearing on the relationship with the ANITA Group. Bot J Linn Soc 146:415–425. doi:10.1111/j.1095-8339.2004.00342.x
Yao CS, Lin M, Wang L (2006) Isolation and biomimetic synthesis of anti-inflammatory stilbenolignans from Gnetum cleistostachyum. Chem Pharm Bull 54:1053–1057
Yao YF, Bruch AA, Mosbrugger V, Li CS (2011) Quantitative reconstruction of Miocene climate patterns and evolution in Southern China based on plant fossils. Palaeogeogr Palaeoclimatol Palaeoecol 304:291–307. doi:10.1016/j.palaeo.2010.04.012
Yi DK, Yang JC, So S, Joo M, Kim DK, Shin CH, Lee YM, Choi K (2015) The complete plastid genome sequence of Abies koreana (Pinaceae: Abietoideae). Mitochondrial DNA 27:2351–2353. doi:10.3109/19401736.2015.1025260
Yule GU (1925) A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Phil Trans R Soc B 213:21–87
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi:10.1101/gr.074492.107
Zhao LC, Wang YF, Liu CJ, Li CS (2004) Climatic implications of fruit and seed assemblage from Miocene of Yunnan, southwestern China. Quatern Int 117:81–89. doi:10.1016/S1040-6182(03)00118-6
Acknowledgments
We thank the curators of the herbaria A, B, BM, BR, E, HITBC, HK, IBK, IBSC, MO, P, PE, PEM, S, U and Stefanie Ickert-Bond (University of Alaska, Fairbanks, USA), Richard Corlett, Bo Pan, Jiantao Yin, Mareike Roeder (Xishuangbanna Tropical Botanical Garden, Chinese Academy of Science, China), Richard Saunders, Chiu Pang, Jenny Lau, Laura Wong (the University of Hong Kong), Xijin Zeng, Haijun Yang (South China Agricultural University, China), Zhuqiu Song, Shixiao Luo (South China Botanical garden, Chinese Academy of Science, China), Nan Deng, Shengqing Shi, Zeping Jiang (Chinese Academy of Forestry, China), my parents Wenpeng Hou and Luolan Song for access to plant material. Among them, we thank again Bo Pan, Jenny Lau, Xijin Zeng for providing photos of Gnetum. We acknowledge support from Science for Life Laboratory, National Genomics Infrastructure (NGI, Sweden) and Uppmax for providing assistance in massive paralleled sequencing and computational infrastructure and Anbar Khodabandeh (Bergius Botanic Garden, Sweden) for technical assistance. We also thank two anonymous reviews for valuable comments on the manuscript. The project was funded by grants from Stockholm University and the Swedish Research Council (Vetenskapsrådet, VR) to C. Rydin, and from Lars Hiertas Memory stipend, Regnells Botanic Memory and P. F. Wahlbergs Memory Stipends from the Swedish Royal Academy of Sciences to C. Hou.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflicts of interest.
Additional information
Handling editor: Martin Lysak.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00606-016-1363-x.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Sequence information for high throughput sequencing (top): we list voucher information, collection places of each specimen, the sizes of assembled plastid genomes, number of matched reads and average coverage during mapping. Information on Sanger sequencing (followed), includes voucher information, collection places of each specimen and GenBank accessions.
Online Resource 2. We list alignments of the four datasets (dataset 1–4) used in the present study.
Online Resource 3. We list 115 genes identified in the plastid genomes of Chinese lianoid Gnetum categorized according to gene function. Other relevant information, such as pseudogenes, number of introns and copies in IRs are also listed.
Online Resource 4. Here we list all lianoid species of Gnetum present in China. The descriptions are based on original descriptions of species, notes in Flora of China (Fu et al. 1999a), and results of the present study including investigations of type material of all listed species. One additional species of Gnetum is present in China, the arborescent G. gnemon. It is variable with many described subspecies and variants and will be dealt with in a forthcoming study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hou, C., Wikström, N., Strijk, J.S. et al. Resolving phylogenetic relationships and species delimitations in closely related gymnosperms using high-throughput NGS, Sanger sequencing and morphology. Plant Syst Evol 302, 1345–1365 (2016). https://doi.org/10.1007/s00606-016-1335-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1335-1