Abstract
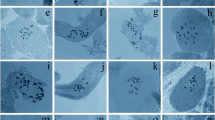
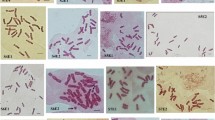
Fluorescent chromosome banding and measurements of nuclear DNA content by image cytometry of Feulgen-stained cells were performed in one sample each of eight diploid (2n = 24) species of Solanum: S. endoadenium, S. argentinum, S. pseudocapsicum, S. atropurpureum, S. elaeagnifolium, S. sisymbriifolium, S. chenopodioides, and S. palustre. The species studied could be distinguished by heterochromatin amount, banding patterns, and genome size. They exhibited only GC-rich heterochromatin and showed a comparatively low heterochromatin amount (expressed as percentage of haplotype karyotype length), ranging from 2.10 in S. argentinum to 8.37 in S. chenopodioides. Genome size displayed significant variation between species, with 1C-values ranging from 0.75 pg (735 Mbp) in S. palustre to 1.79 pg (1,754 Mbp) in S. sisymbriifolium. No significant correlation between genome size and heterochromatin amount was observed, but intrachromosomal asymmetry index (A 1) was negative and significantly correlated with heterochromatin amount. DNA content was positively and significantly correlated with karyotype length. DNA C-value distribution in the genus as well as karyotype affinities and relationships between species are discussed in relation to different infrageneric classifications of Solanum.






Similar content being viewed by others
References
Acosta MC, Bernardello G, Guerra M, Moscone EA (2005) Karyotype analysis in several South American species of Solanum and Lycianthes rantonnei (Solanaceae). Taxon 54:713–723
Barboza GE, Cantero JJ, Nuñez CO, Ariza Espinar L (2006) Flora Medicinal de la Provincia de Córdoba (Argentina). Pteridófitas y Antófitas silvestres o naturalizadas, Museo Botánico, Córdoba, Argentina
Barros e Silva AE, Guerra M (2010) The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech Histochem 85:115–125
Bennett MD, Leitch IJ (1995) Nuclear DNA amounts in angiosperms. Ann Bot (Lond) 76:113–176
Bennett MD, Leitch IJ (2010) Angiosperm DNA C-values database (release 7.0, Dec. 2010), http://www.rbgkew.org.uk/cval/homepage.html (November 17, 2011)
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot (Lond) 107:467–590
Bennett MD, Smith JB (1991) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci 334:309–345
Bernardello LM, Anderson GJ (1990) Karyotypic studies in Solanum section Basarthrum (Solanaceae). Am J Bot 77:420–431
Bernardello LM, Heiser CB, Piazzano M (1994) Karyotypic studies in Solanum section Lasiocarpa (Solanaceae). Am J Bot 81:95–103
Bohs L (2005) Major clades in Solanum based on ndhF sequence data. In: Keating RC, Hollowell VC, Croat TB (eds). A festschrift for William G. D’Arcy: the legacy of a taxonomist. Monogr Syst Bot Missouri Bot Gard 104. Missouri Botanical Garden Press, St. Louis, pp 27–49
Brasileiro-Vidal AC, Melo-Oliveira MB, Carvalheira GMG, Guerra M (2009) Different chromatin fractions of tomato (Solanum lycopersicum L.) and related species. Micron 40:851–859
Chiarini F, Bernardello G (2006) Karyotype studies in South American species of Solanum subgenus. Leptostemonum (Solanaceae). Plant Biol 8:486–493
D’Arcy WG (1972) Solanaceae studies II: typification of subdivisions of Solanum. Ann Missouri Bot Gard 59:262–278
de Almeida CS, de Carvalho PL, Guerra M (2007) Karyotype differentiation among Spondias species and the putative hybrid Umbu-cajá (Anacardiaceae). Bot J Linn Soc 155:541–547
Greilhuber J, Ebert I (1994) Genome size variation in Pisum sativum. Genome 37:646–655
Greilhuber J, Speta F (1976) C-banded karyotypes in the Scilla hohenackeri group, S. persica, and Puschkinia (Liliaceae). Plant Syst Evol 126:149–188
Guerra M (2009) Chromosomal variability and the origin of Citrus species. In: Mahoney CL, Springer DA (eds) Genetic diversity. Nova Science, New York, pp 51–68
Hawkes JG (1999) The economic importance of the family Solanaceae. In: Nee M, Symon DE, Lester RN, Jessop JP (eds) Solanaceae IV: advances in biology and utilization. Royal Botanic Gardens, Kew, pp 1–8
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66:18–33
Hunziker AT (2001) Genera Solanacearum: the genera of Solanaceae illustrated, arranged according to a new system. ARG Gantner Verlag K-G, Liechtenstein
Infostat Group (2002) INFOSTAT version 1.1. Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba. Editorial Brujas, Argentina
Levan A, Fredga K, Sandberg A (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
Melo CAF, Martins MIG, Oliveira MBM, Benko-Iseppon AM, Carvalho R (2011) Karyotype analysis for diploid and polyploid species of the Solanum L. Plant Syst Evol 293:227–235
Miguel V, Acosta MC, Moscone EA (2012) Karyotype analysis in two species of Solanum (Solanaceae) Sect. Cyphomandropsis based on chromosome banding. New Zeal J Bot 50:217–225
Moscone EA (1992) Estudios de cromosomas meióticos en Solanaceae de Argentina. Darwiniana 31:261–297
Moscone EA, Baranyi M, Ebert I, Greilhuber J, Ehrendorfer F, Hunziker AT (2003) Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Ann Bot (Lond) 92:21–29
PBI Solanum Project (2012) Solanaceae Source. http://www.nhm.ac.uk/research-curation/research/projects/solanaceaesource (January 31, 2012)
Pijnacker LP, Ferwerda MA (1984) Giemsa C-banding of potato chromosomes. Can J Genet Cytol 26:415–419
Pringle GJ, Murray BG (1991) Karyotype diversity and nuclear DNA variation in Cyphomandra. In: Hawkes JG, Lester RN, Nee M, Estrada N (eds) Solanaceae III: taxonomy, chemistry, evolution. Royal Botanic Gardens Kew and Linnean Society of London, London, pp 247–252
Pringle GJ, Murray BG (1993) Karyotypes and C-banding patterns in species of Cyphomandra Mart. ex Sendtner (Solanaceae). Bot J Linn Soc 111:331–342
Rego LNAA, da Silva CRM, Torezan JMD, Gaeta ML, Vanzela ALL (2009) Cytotaxonomical study in Brazilian species of Solanum, Lycianthes and Vassobia (Solanaceae). Plant Syst Evol 279:93–102
Sokal RR, Rohlf FJ (1995) Biometry, the principles and practice of statistics in biological research, 3rd edn. W.H. Freeman, New York
Sultana SS, Alam SS (2007) Differential fluorescent chromosome banding of Solanum nigrum L. and Solanum villosum L. from Bangladesh. Cytologia 72:213–219
Valkonen JPT (1994) Nuclear DNA content of the Solanum spp. in the series Etuberosa as determined by laser flow cytometry. Ann Appl Biol 125:589–600. [Not seen; data from: Bennett MD and Leitch IJ (2010) Angiosperm DNA C-values database (release 7.0, Dec. 2010), http://www.rbgkew.org.uk/cval/homepage.html]
Weese TL, Bohs L (2007) A three-gene phylogeny of the genus Solanum (Solanaceae). Syst Bot 32:445–463
Wu F, Tanksley SD (2010) Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11:182
Zhu W, Ouyang S, Iovene M, O’Brien K, Vuong H, Jiang J, Buell CR (2008) Analysis of 90 Mb of the potato genome reveals conservation of gene structures and order with tomato but divergence in repetitive sequence composition. BMC Genomics 9:286
Acknowledgments
We thank Dr. G.E. Barboza and Dr. M. Matesevach for identification of voucher specimens, and several botanists who provided material. Dr. A.C. Brasileiro-Vidal supplied valuable assistance in the laboratory. Thanks to the anonymous reviewer for suggestions that improved the paper. M.C. Acosta acknowledges fellowship support from the Latin-American Net of Botany for a study leave at the Department of Botany, Federal University of Pernambuco (Recife, Brazil). This work was supported by grants from the National University of Córdoba (SECYT-UNC, Argentina) and the National Council of Scientific and Technical Researches (CONICET, Argentina) to E.A. Moscone.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Acosta, M.C., Guerra, M. & Moscone, E.A. Karyological relationships among some South American species of Solanum (Solanaceae) based on fluorochrome banding and nuclear DNA amount. Plant Syst Evol 298, 1547–1556 (2012). https://doi.org/10.1007/s00606-012-0657-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0657-x