Abstract
Pistachio is economically important in Iran. Selection of suitable rootstocks, resistant to unfavorable and soil conditions and diseases, is important for increasing yield and the acreage of this crop. It is essential to identify the genetic relationships among Pistacia species for the breeding of pistachio rootstocks. The goal of this study was to determine the genetic relationship among Pistacia species (P. vera L., P. khinjuk Stocks., P. eurycarpa Yalt., P. atlantica subsp. atlantica Zoh., P. atlantica subsp. mutica Fish et C.A. Mey and P. atlantica subsp. cabulica Stocks.) with potential in the breeding of rootstocks using the selective amplification of microsatellite polymorphic loci (SAMPL) technique. Six primer combinations produced a total of 182 bands, with an average of 30.33 bands per primer pair, of which 128 were polymorphic. Three branches were obtained, the first containing P. vera, and the second containing P. khinjuk, P. eurycarpa, P. atlantica and subspecies mutica and cabulica, with numerous leaflets clustered in the third branch. UPGMA analysis separated P. atlantica subspecies from P. eurycarpa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Pistacia includes 13 or more species, and of these Pistacia vera L. has commercially important edible nuts. The other species grow in the wild and their seedlings are used mainly as rootstocks for pistachio (Kafkas and Perl-Treves 2002). There are two main centers of diversity for Pistacia: one comprises the Mediterranean region of Europe, Northern Africa and Middle East countries. Other is the Eastern part of Zagros Mountains (Iran) and Caucasus regions from Crimea to the Caspian Sea (Zohary 1952). Iran is a center of origin for four important Pistacia species, P. vera L., P. khinjuk Stocks. P. eurycarpa Yalt. (P. atlantica subsp. kurdica Zoh.) and P. atlantica Dsef. (Karimi et al. 2009b).
Pistachio is usually propagated by budding or grafting selected scions onto seedling rootstocks of the same species or other Pistacia species. Different rootstocks are used in the different growing areas; thus, P. atlantica, P. integerrima (Stewart) Zoh. and their hybrids are the main rootstocks in California. Pistacia vera seedlings are main rootstocks used in Iran and Turkey. Resistance to both aerial (salinity and drought) and soil fungal diseases (Phytophthora, Verticillium and nematodes) are interesting traits in the breeding of pistachio rootstocks. Pistacia mutica and P. khinjuk are the rootstocks most resistant to root-knot nematode and drought, respectively. Pistacia species have a high genetic diversity due to their dioecious character, pollination mechanism and high heterozygosity. Because of these factors high selectivity in rootstocks breeding is required, and therefore knowledge of the genetic relationships among Pistacia species would be very useful in pistachio rootstock breeding. Different markers have been used for studying Pistacia species and pistachio cultivars (Karimi et al. 2009a; Kafkas 2006; Ahmad et al. 2003; Katsiotis et al. 2003; Parfitt and Badenes 1997), but no study has been reported in which the SAMPL (selective amplification of microsatellite polymorphic loci) marker technique was used. SAMPL is a modified amplified fragment length polymorphism (AFLP) technique in which a compound microsatellite sequence is used as one of the two AFLP primers in selective amplification (Witssenboer et al. 1997), generally in place of EcoRI primers.
The objective of this study was to assess the genetic relationships among Pistacia species using the SAMPL marker technique.
Materials and methods
We used 28 wild and cultivated pistachios accessions in this study. The accessions belonged to P. vera, P. khinjuk, P. eurycarpa, P. atlantica subsp. atlantica, P. atlantica subsp. mutica and P. atlantica subsp. cabulica. Samples were originally from Iran, Turkey, US and Syria (Table 1). Leaf samples were used for DNA extraction, and genomic DNA was extracted using the CTAB method of Doyle and Doyle (1987) with minor modifications (Kafkas and Perl-Treves 2002). Leaf samples (1 g) were ground in liquid nitrogen and mixed with 6 ml of CTAB buffer (100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 2% polyvinylpyrrolidone, 0.2% β-mercaptoethanol, 0.1% Na2S2O5). The samples were then incubated at 65°C for 1 h, followed by extraction with an equal volume of chloroform/isoamyl alcohol (24:1). The aqueous phase was recovered and mixed with an equal volume of cold isopropanol and left at −20°C for 24 h. The precipitated nucleic acids were recovered by centrifugation at 1,000 rpm for 2 min, washed with ammonium acetate (10 mM) in 76% ethanol, dried and resuspended in double-distilled water. The DNA concentration was estimated by comparison with known DNA concentrations, after 0.8% agarose gel electrophoresis and ethidium bromide staining, and the concentration was adjusted to 50 ng/μl for the SAMPL procedure.
For the SAMPL reaction, 50 ng of genomic DNA was used. Restriction and ligation were performed using 5 U of EcoRI and 1 U of MseI enzymes, 5 pmol of each EcoRI and MseI adaptor, 1 U of T4 DNA ligase, 1× ligase buffer, 1.1 μl of 0.5 M NaCl and 0.55 μl of bovine serum albumin (1 ng/μl), in a final volume of 11 μl. The preselective amplification reaction mixture contained 4 μl restricted-ligated DNA as PCR template, 5 pmol of each preselective amplification primer (EcoRI +A and MseI +C), 3 mM dNTPs, 2 μl PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl), 0.4 mM MgCl2 and 1 U of Taq DNA polymerase. The thermocycle conditions were as described by Vos et al. (1995) with minor modifications (Kafkas 2006). The adaptor sequences, preselective amplification primers and selective primers are listed in Table 2.
The selective amplification reaction was conducted in a final volume of 20 μl containing 4 μl of diluted preselective amplification products as template, 5 pmol each of labeled EcoRI site primer (SAMPL-6 or SAMPL-7) and MseI primer, 3 mM dNTPs, 1X PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl), 3 mM MgCl2 and 1 U Taq DNA polymerase. The SAMPL program included one cycle of 2 min at 94°C, followed by ten cycles of denaturation at 94°C for 45 s, gradient annealing at 65°C (−1°C per cycle) for 45 s, and extension at 72°C for 2 min. An additional 20 cycles of denaturation, annealing and extension at 94°C for 45 s, 56°C for 45 s and 72°C for 2 min were applied, followed by a final incubation for 30 s at 60°C. The PCR products were separated on an ABI Prism 3130 Genetic Analyzer. The polymorphic bands were scored as present (1) or absent (0). A Jaccard's similarity matrix was calculated using numerical taxonomy and the multivariate analysis system NTSYSpc ver 2.11 (Rohlf 2004) and the dendrogram produced using the unweighted pair-group method with arithmetic mean (UPGMA) algorithm.
Results
SAMPL polymorphism and discrimination capacity
Six SAMPL primer combinations were used in the molecular characterization of 28 pistachio accessions originating in different countries. These combination primers generated in total 182 scorable bands, with an average of 30.33 bands per primer pair, of which 128 were polymorphic. The number of total bands varied from 20 (SAMPL-6/MCAC) to 48 (SAMPL7/MCCA). The maximum number of polymorphic bands was 37 for primer combination SAMPL-7/MCCA and the lowest number was 13 for SAMPL-6/MCAG (Table 3).
Genetic relationships among Pistacia accessions
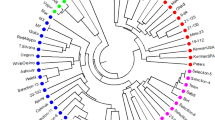
According to Jacquard’s similarity matrix (Table 4) and UPGMA clustering, accessions at a similarity distance of 0.55 were separated into three main branches (Fig. 1). The first branch included P. vera that comprised two groups: the first group comprised pistachio accessions originating in Turkey, and the second group comprised accessions originating in Iran. In the Turkish group, the ‘Siirt’ cultivar was separated by the greatest distance from the others. In the Iranian group, the ‘Badami’ cultivar was more similar to the ‘Ghazvini’ cultivar, whereas the ‘Sarakhs’ variety was separated by the greatest distance from the others. The second branch included P. khinjuk accessions from Iran and the US. In this cluster Iranian samples were separated from US samples. Pistacia eurycarpa and all of the subspecies of P. atlantica (atlantica, mutica and cabulica) were located in the third branch. This branch included three groups: The first group comprised P. eurycarpa accessions from Turkey; the second group comprised P. atlantica subsp. atlantica from Turkey and Syria plus P. atlantica subsp. mutica from Turkey; and the third group comprised P. atlantica subsp. mutica and cabulica from Iran. P. khinjuk was more similar to P. eurycarpa than P. vera, and P. eurycarpa was most similar to P. atlantica.
Discussion
Phylogenetic analysis of related plant species can be performed using morphological descriptors and molecular markers. The use of DNA markers is one of the most powerful techniques, because they are not influenced by environmental factors and the development stage of the plant. The SAMPL marker technique is a powerful technique for the analysis of intraspecific diversity in different fruit and nut tree species. One of the main advantages of this marker is a high multiplex ratio as in AFLP, and it has some degree of locus specificity as in simple sequence repeats (SSRs), thus making it a very efficient and effective molecular markers system. In present study the SAMPL marker technique was used to study the genetic relationships among Pistacia species. Six combination primers generated a total 182 bands with an average of 30.33 bands per primer pair, of which 128 were polymorphic. In a similar study, Kafkas et al. (2005) found 50.9% polymorphism in the walnut using the SAMPL marker technique, Therefore the SAMPL marker can separate closely related accessions.
In this study P. khinjuk was more similar to P. eurycarpa than to P. vera, also there were similar genetic relationships between P. vera and P. khinjuk and P. eurycarpa. Similar studies (Karimi et al. 2009b, Kafkas et al. 2002) have shown that the closest species to P. vera is P. eurycarpa, followed by P. atlantica. Zohary (1972) found that P. eurycarpa, which has larger fruits and ovate and few paired leaflets, closely resembles to P. vera. This species is centered mainly in Iran and Afghanistan, overlapping in some places with P. vera and P. khinjuk. Yaltirik (1967) described and introduced P. eurycarpa as a new species.
In contrast to the findings of Karimi et al. (2009b), this study showed that P. atlantica and P. atlantica subsp. mutica were very similar and should be considered the same species. The findings of the present study strongly support those of Al-Yafi (1978) and Kafkas (2006) who postulated that the mutica subspecies should be located in P. atlantica.
There is a hypothesis that P. eurycarpa is a hybrid between P. vera and P. khinjuk, and P. atlantica and mutica are derivatives of it (Zohary 1972). According to previous studies and the present study, P. khinjuk and P. eurycarpa have similar genetic distances to P. vera, while P. eurycarpa is more similar to P. atlantica than to P. khinjuk, and it is possible that P. eurycarpa is a hybrid between P. vera and P. khinjuk, and P. atlantica and the mutica subspecies are derivatives of it.
In conclusion, the SAMPL marker technique was able to easily divide accessions with regard to the species P. vera, P. khinjuk, P. eurycarpa and P. atlantica, and the SAMPL marker technique may also be used to reveal the genetic relationships among Pistacia species for better crossing of tolerant species for rootstock production. This study showed that P. atlantica and P. atlantica subsp. mutica should be considered same species. According to previous studies (Parfitt and Badenes 1997), P. vera and P. khinjuk are the most similar, while in this study P. khinjuk appeared to be more similar to P. eurycarpa. Following this study we can postulate that P. eurycarpa may be a hybrid between P. vera and P. khinjuk, and that P. atlantica, P. atlantica subsp. mutica and P. atlantica subsp. cabulica are descendents of P. eurycarpa. Therefore, the use of more efficient and powerful markers such as SSR and RFLP is recommended for future research.
References
Ahmad R, Ferguson L, Southwick SM (2003) Identification of Pistachio (Pistacia vera L.) nuts with microsatellite markers. J Am Soc Hortic Sci 128:898–903
Al-Yafi J (1978) New characters differentiating Pistacia atlantica subspecies. Candollea 33:201–208
Behboodi B (2003) Ecological distribution study of wild pistachios for selection of rootstock. Options Mediterraneennes Ser A 63:61–67
Doyle JJ, Doyle JL (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Golan-Goldhirsh A, Barazani O, Wang ZS, Khadka DK, Saunders JA, Kostiukovsky V, Rowland LJ (2004) Genetic relationships among Mediterranean Pistacia species evaluated by RAPD and AFLP markers. Plant Syst Evol 246:9–18
Hormaza JL, Polito VS (1994) Identification of relatedness and geographic movement of Pistacia vera germplasm by RAPD analysis. Econ Bot 48:349–358
Kafkas S (2006) Phylogenetic analysis of the genus Pistacia by AFLP markers. Plant Syst Evol 262:113–124
Kafkas S, Perl-Treves R (2002) Interspecific relationship in the genus Pistacia L (Anacardiaceae) based on RAPD fingerprinting. J Hort Sci 37:168–171
Kafkas S, Ebru K, Perl-Treves R (2002) Morphological diversity and germplasm survey of three wild Pistacia species in Turkey. Genet Resour Crop Ev 49:261–270
Kafkas S, Ozkan H, Sutvemez M (2005) DNA polymorphism and assessment of genetic relationships in Walnut genotypes based on AFLP AND SAMPL markers. J Am Soc Hortic Sci 130:585–590
Kafkas S, Ozkan H, Ak BE, Acar I, Atli HS, Koyuncu S (2006) Detecting DNA polymorphism and genetic diversity in a wide pistachio germplasm: comparison of AFLP, ISSR and PAPD markers. J Am Soc Hortic Sci 131:522–529
Karimi HR, Kafkas S, Zamani Z, Ebadi A, Fatahi MR (2009a) Genetic relationships among Pistacia species using AFLP markers. Plant Syst Evol 279:21–28
Karimi HR, Zamani Z, Ebadi A, Fatahi MR (2009b) Morphological diversity of Pistacia species in Iran. Genet Resour Crop Ev 56:561–571
Katsiotis A, Hagidimitriou M, Drossou A, Partikis C, Loukas M (2003) Genetic relationships among species and cultivars of Pistacia using RAPDs and AFLPs. Euphytica 132:279–286
Louska M, Pontikis CA (1979) Pollen isozyme polymorphism in types of Pistacia vera and related species as aid in taxonomy. J Hort Sci 54:95–102
Parfitt D, Badenes ML (1997) Phylogeny of the genus Pistacia as determined from analysis of the chloroplast genome. Proc Natl Acad Sci U S A 94:7987–7992
Rechinger KH (1969) Anacardiaceae. In: Rechinger KH (ed) Flora Iranica, vol 63. Akademische Druck- und Verlagsanstalt, Graz, pp 1–9
Riaz A, Ferguson L (2003) Identification of pistachio nuts with microsatellite markers. J Hort Sci Biot 128:893–903
Riaz A, Ferguson L (2005) Molecular marker analysis of pistachio rootstocks by simple sequence repeats and sequence related amplified polymorphism. J Hort Sci Biot 80:382–386
Rohlf JF (2004) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.11. Exeter Publishing, Setauket
Vos P, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Witssenboer H, Vogel J, Michelmore RW (1997) Identification genetic localization and allelic diversity of selectively amplified microsatellite polymorphic loci in lettuce and wild relatives (Lactuca spp.). Genome 40:923–936
Yaltirik F (1967) Anacardiaceae. In: Davis PH (ed) Flora of Turkey and the East Aegean island, vol 2. Edinburgh University Press, Edinburgh. pp 544–548
Zohary M (1952) A monographical study of the genus Pistacia. Palest J Bot Jerus Ser 5:187–228
Zohary M (1972) Pistacia L. Flora Palestine. Israel Acad Scie Humanit, Jerus 2:187–228
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Karimi, H.R., Kafkas, S. Genetic relationships among Pistacia species studied by SAMPL markers. Plant Syst Evol 297, 207–212 (2011). https://doi.org/10.1007/s00606-011-0508-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-011-0508-1