Abstract
This paper reports the glutenin diversity of a collection of 23 wild populations of the grass Brachypodium distachyon collected in the Mediterranean and southern areas of the Iberian Peninsula. The plant material studied included the three different cytotypes of this species: 2n = 10, 2n = 20 and 2n = 30. A specific method of extraction was used to isolate the glutenin subunits from the caryopsis. Separation by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) showed them to correspond to wheat low-molecular-weight glutenin subunits (LMW-GS). Twenty-two LMW-GS–like were identified that showed great diversity within and between populations. All the populations investigated were polymorphic for the endosperm proteins studied. The 2n = 30 forms had the largest number of subunits; these were also more diverse than those of the 2n = 10 or 2n = 20 forms. The 2n = 10 forms, the most common in the higher, interior areas of the Iberian Peninsula, showed the smallest subunit variation. In fact, negative correlations were found between subunit diversity and altitude and longitude. In contrast, a positive correlation was detected with the annual average temperature and the indices of thermicity and Mediterraneity. The similarity between the populations was estimated using the Sorensen–Dice coefficient, calculated on the basis of the presence/absence of the 22 LMW-GS proteins. The similarity indices were used to produce a dendrogram using the unweighted pair-group method with arithmetic means (UPGMA). This method produced three main groups corresponding to the three cytotypes. An analysis was made of the possible correlation between eco-geographic and climatic factors and the gene diversity and polymorphism shown in the populations. The diversity correlated positively with the number of chromosomes (2n), annual mean temperature, index of thermicity and index of Mediterraneity. In contrast, diversity correlated negatively with altitude, longitude and index of precipitation during the summer. The number of chromosomes correlated negatively with altitude, longitude and precipitation during summer and positively with all the other climatic indices. These results are coherent with the fact that diploid forms were more common in areas of the interior and at higher altitude, where the climate is more extreme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brachypodium distachyon (L.) P. Beauv., a self-pollinating species, is fast emerging as a powerful model for studying the grass family, including wheat and other temperate grasses. As a model it offers several attractive attributes such as a small (~300 Mbp), recently sequenced genome (International Brachypodium Initiative, 2010) and a short lifecycle (Higgins et al. 2010). In addition it can be easily transformed.
Several investigations have shown Brachypodium to be closely related to wheat and other small grain cereals such as barley and oat (Catalan et al. 1997; Catalan and Olmstead 2000). The close phylogenetic relationship between Brachypodium and wheat is also reflected in the strong similarity of their gene sequences (Ozdemir et al. 2008). Indeed, there is a general coincidence in the order of Brachypodium genes and those of the other species of the tribe Triticeae, while the genome size is similar to that of rice. Brachypodium is therefore seen as a “bridge” between rice and the Triticeae tribe. Various molecular phylogenetic analyses have shown that Brachypodium diverged from the ancestral stock of Pooideae immediately prior to the radiation of the modern “core pooids” (Catalan et al. 1997; Catalan and Olmstead 2000; Draper et al. 2001). It is also ancestral, but equally related to, Avena and Triticum (Kellogg 2001; Huo et al. 2009).
The species of the genus Brachypodium have small chromosomes of variable number (x = 5, 7, 8, 9 or 10), making them unusual among the members of the Pooideae, which tend to have large chromosomes and base number of 7 (Shi et al. 1993; Draper et al. 2001; Hasterok et al. 2004, 2006; Idziak and Hasterok 2008; Garvin et al. 2008). Although ecotypes of B. distachyon with 10, 20 and 30 chromosomes have been described, some authors have indicated that these numbers do not represent a simple autopolyploid series (Robertson 1981). The 2n = 10 and 2n = 20 ecotypes appear to be diploids, and it is generally assumed that the 2n = 30 forms must be allotetraploids produced by the addition of the 2n = 10 and 2n = 20 genomes (Hasterok et al. 2004; Filiz et al. 2009). The 2n = 20 and 2n = 30 forms of Brachypodium are easily distinguished by visual examination from the 2n = 10 form (Vogel et al. 2009).
B. distachyon has a rapid generation cycle under optimized conditions (8–12 weeks), does not grow very tall (20–30 cm) at high planting densities, requires few demanding growth resources, shows good environmental adaptation (Vogel et al. 2009) and the 2n = 10 forms do not require vernalization (Draper et al. 2001; Ozdemir et al. 2008; Vogel et al. 2006). The species B. distachyon has been used to protect rural soils from erosion, especially in slope areas where water runoff, wind and other agents cause soil loss. Recently, some varieties of B. distachyon have been marketed in Spain to protect olive groves from erosion (Soler et al. 2004).
Despite the substantial body of knowledge now accumulated on Brachypodium, little is known about the biochemical composition and variability of its grain. Brachypodium seeds are typical of the Poaceae family with caryopses smaller than all other grass species (Opanowicz et al. 2008). The major grain components are seed storage proteins, which account for about 60–80% of total protein load, depending on species and variety. When classified according to their solubility, the grain storage proteins of cereals fall into three fractions: water-soluble albumins, dilute saline-soluble globulins and prolamins that are soluble in alcohol/water solutions under reducing conditions (Shewry and Tathan 1995). Laudencia-Chingcuanco and Vensel (2008) studied the grain filling process and seed storage proteins of B. distachyon grains and reported globulins to be the major protein components. Recently, Larré et al. (2010) showed that B. distachyon lacks albumins; globulins and prolamins are the main seed storage proteins. The globulins are represented mainly by the 11S type, their solubility properties being very similar to the glutelin found in rice. However, the study of Larré was restricted to a single diploid inbred line isolated from the original accession Bd21 by Vogel and Hill (2008). More recently, Wang et al. (2010) studied the composition of the grain proteins of 13 B. distachyon accessions (three with 2n = 10, and ten with 2n = 30) and reported the major storage proteins in these plants to be wheat low-molecular-weight glutenin subunit-like (LMW-GS-like proteins) and globulins. A few high-molecular-weight glutenin subunits (HMW-GS) produced at low expression levels are also present. For a deeper knowledge of the storage proteins of B. distachyon to be gained, however, more materials need to be studied, analyzing the degree of diversity of the storage proteins within and between different wild populations, and examining the relationship of this diversity with the number of chromosomes and the ecological, geographical and climatic conditions of the places where samples are collected.
The increasing interest in the genetic resources of Brachypodium has led to several germplasm collections coming into existence (Doust 2007; Huo et al. 2008; Vain et al. 2008; Vogel and Hill 2008; Garvin 2007). Knowledge of the genetic diversity in wild populations of B. distachyon is central to the development of its effective conservation in genebanks and the use of this primitive grass in breeding strategies. One of the present authors (C.S.) undertook several expeditions to different regions of the Iberian Peninsula and produced a good collection of natural populations of B. distachyon. A group of these populations, selected by their differences in terms of eco-geographic origin and number of chromosomes, formed the material examined in the present work.
The aim of this study is to characterize the genetic variability of the endosperm LMW-GS-like proteins in a collection of wild populations of B. distachyon gathered in Spain, using two marketed varieties as controls. The samples examined belong to all three cytotypes (2n = 10, 2n = 20, 2n = 30). The above proteins were characterized and an analysis made of their natural diversity in relation to chromosome number and eco-geographic variables.
Materials and methods
Plant material
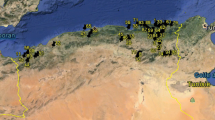
Spain is remarkably rich in wild populations of this species. In recent years (2000–2008) a number of expeditions were undertaken to collect samples from wild populations in the southern and Mediterranean areas of the Iberian Peninsula. To preserve the original structure of the natural populations, specimens were collected at random following the method of Hawkes (1980). A total of 23 wild populations of B. distachyon from these different geographic regions were used as plant material in this work. Two commercial varieties, ‘Ibros’ (=Bd3107; 2n = 30) and ‘Zulema’ (=Bd3113; 2n = 10), were used as controls. These were obtained via domestication of natural populations, selecting uniform lines over several generations to produce a turf for protection of degraded soils (Soler et al. 2004). Figure 1 shows the geographical location of each accession used in the present work, while Table 1 records their chromosome number and main eco-geographic features. The accessions studied form part of a collection held at the Unit of Plant Improvement of the Department of Ecology of the Instituto Nacional de Investigaciones Agrarias (INIA) at the La Canaleja Research Station (Alcalá de Henares, Madrid, Spain). Climatic data were collected at each collection site from the closest meteorological station. These included the annual average temperature, the Rivas-Martinez thermicity index (Rivas-Martínez et al. 1990), i.e. It = (T + m + M)/10, annual precipitation in mm (P) and the index of Mediterraneity (IM3 = PET June + July + August/P June + July + August > 2.5, where PET is the monthly potential evapotranspiration of Thornthwaite (1948).
In December 2007, seeds from each accession of B. distachyon and both commercial lines were planted in individual 15-cm-diameter plastic pots. Plants were grown under greenhouse conditions until March. All plants were then transferred outside the greenhouse until ripening. At maturity, ears from each plant were bagged to avoid mixture of seeds, and all ears were harvested. Flour was then made from 10 seeds per plant for 12 plants from each of the above materials. Caryopses were independently ground in a mortar. The seeds of Brachypodium are very hard, so liquid nitrogen was used to facilitate this process. The flour produced was left for 24 h in an oven at 37°C before extracting the proteins.
SDS-PAGE analysis
To identify the major seed storage proteins, differential extraction of the total proteins was undertaken followed by denaturing one-dimensional (1D) SDS-PAGE. The differential extraction method used was a modification of that described by Singh et al. (1991). This modification allowed the gliadins and glutenins to be separated; this work, however, only reports the analysis of the glutenins.
Two hundred microlitres of solution A (propanol-1 50% v/v) was added to 20 mg flour extracted from the 10 seeds taken from single plants. These samples were shaken and incubated for 1 h in a water bath at 65°C before centrifuging at 10,000×g for 15 min. The supernatant was eliminated, and 200 µl solution A was added to the precipitate. The resulting samples were incubated for 30 min in a water bath at 65°C, centrifuged at 12,000×g for 5 min and again the supernatant eliminated. Twenty microlitres of solution B (propanol-1 50% v/v, mixed with 1 M Tris–HCl; pH 8) and 1% v/v dithiothreitol (DTT) were added to the pellet to purify the glutenins. The resulting samples were then shaken and incubated for 30 min in a water bath at 65°C. Solution B (20 μl) and 4-vinylpiridine to 1.4% v/v were then added and the samples shaken and incubated for 15 min in a water bath at 65°C. Finally, the samples were centrifuged at 10,000×g for 2 min, and the supernatant, corresponding to the glutenin fraction, was recovered. Twenty microlitres of the supernatant was then mixed with 20 μl solution C (0.2 g SDS, 4 ml glycerol, 2 mg bromophenol blue, 0.8 ml Tris–HCl 1 M; pH = 8) to produce samples for electrophoretic analysis.
The protein samples of the 12 individual plants from each of the studied materials were separated on SDS-PAGE gels. Multimark proteins (Invitrogen) were used as molecular weight markers. Triticum aestivum cv. ‘Chinese Spring’ was used to provide a reference glutenin pattern. The SDS-PAGE method followed required the preparation of polyacrylamide gels with two different concentration areas: 3% acrylamide in the stacking area, and 12% in the separation zone. A constant voltage of 20 mV was applied to the stacking area. This was increased to 40 mV when the visualization medium approached the start of the separation zone. The gel was stained with a mixture of Coomassie blue G, Coomassie blue R, acetic acid, ethanol and trichloroacetic acid. The gels were washed with distilled water to eliminate excess dye, photographed and the images digitized.
Study of gels and data analysis
The total glutenins of the studied materials were examined independently. Following electrophoretic separation, the glutenin-like proteins were analyzed. Only LMW-GS bands were analyzed in the present study, using T. aestivum cv. ‘Chinese Spring’ as control. Variability was investigated by analysis of the presence/absence of each band in the electrophoretic profiles. A total of 22 bands were identified; these were numbered from 1 to 18A from top to bottom in the gels (bands 9, 17 and 18, which initially were considered as single bands, were later subdivided into two: 9 and 9A, 17 and 17A, 18 and 18A, due to perceivable differences of migration; 17 and 17A, 18 and 18A never coincided in the same plant). The presence/absence of each glutenin, the frequency of each in the set of plants analyzed per population, the different electrophoretic profiles and the pattern for each accession were recorded. A data matrix using “1” for presence and “0” for absence of each glutenin was constructed. A second matrix for the frequency of each band was also produced.
Factorial analysis of correspondences was used to examine the inter-population distances and the influence of each variable (proteins, eco-geographic factors and chromosome number) on the grouping of populations.
The similarity between the populations was reflected using the Sorensen–Dice coefficient (Sorensen 1948) calculated from the binary data (presence/absence) matrix for the 22 proteins. These similarity indices were used to produce a dendrogram using the unweighted pair-group method with arithmetic means (UPGMA) (Sneath and Sokal 1973). The NTSYS package was used for all analyses (Rolf 1994).
Analysis of within- and among-populations LMW-GS diversities were conducted using Nei’s index of heterogeneity. The average diversity for all the LMW-GS studied was estimated considering the frequencies of each band in all loci in the 23 natural populations analyzed.
Chromosome preparations
Seeds of plants from each wild population and the commercial lines of B. distachyon were germinated on moist filter paper for 24 h at 25°C. They were then left at 4°C for 72 h and returned to a temperature of 25°C for 24 h in order to synchronize cell divisions in the root tips. The roots were excised and pre-treated in cold water for 24 h before fixation in ethanol/acetic acid (3:1). Before squashing, the pretreated roots were fixed in a solution of 3:1 ethanol/acetic acid, washed and transferred to an enzyme solution containing 2% cellulose and 20% pectinase. Chromosome preparations were made by squashing the meristem of the root in a drop of 45% acetic acid on a clean microscope slide. After removal of the coverslips by freezing, the slides were air-dried, stained with the DNA-binding fluorochrome 4′,6-diamidino-2-phenylindole (DAPI), and examined using a Zeiss Axiophot epifluorescence microscope.
FISH, GISH, probes and labelling
The DNA probes used in these analyses were pTa71, a 9-kb fragment from T. aestivum containing the 18S ribosomal DNA (rDNA) sequence including the intergenic spacers (Gerlach and Bedbrook 1979), and pTa794 (a 410-bp fragment also from wheat containing the 5S rDNA-gene repeat unit) (Gerlach and Dyer 1980). Genomic in situ hybridization (GISH) was used to characterize the type of DNA present in the 2n = 30 forms. For this, DNA from both 2n = 10 and 2n = 20 plants were used as probes. DNA was isolated from B. distachyon Bd3113 (2n = 10) and Bd129 (2n = 20) using the DNeasy Plant extraction kit (Qiagen). Probes were labelled with digoxigenin or biotin by nick translation (pTa71 and genomic DNA) and polymerase chain reaction (PCR) (pTa794). The digoxigenin and biotin were detected using anti-digoxigenin fluorescein isothiocyanate (FITC) conjugate (green) and streptavidin-Cy3 (red), respectively.
Preparations and probe denaturation, in situ hybridization, post-hybridization washing, and detection were all performed according to Cuadrado et al. (2004).
Fluorescence microscopy and imaging
Slides were examined using a Zeiss Axiophot epifluorescence microscope. The separate images from each filter set were captured using a cooled charge-coupled device (CCD) camera (Nikon DS). Images were optimized for contrast and brightness using Adobe Photoshop.
Results
The chromosome number of the plants belonging to each population was initially analyzed by DAPI staining. This allowed the populations to be classified into the three known cytotypes which have 10, 20 or 30 chromosomes (Table 1). The 2n = 10 cytotype had three pairs of distinguishable chromosomes and another two smaller, indistinguishable pairs. The chromosomes of the 2n = 20 cytotype were of similar size and indistinguishable, but smaller than those of the 2n = 10 forms. Finally, the 2n = 30 cytotype seemed to have two groups of chromosomes similar to those of the 2n = 10 plus 2n = 20 cytotypes.
Of the 23 populations analyzed, 8 were of the 2n = 10 cytotype (Bd160, Bd162, Bd169, Bd700, Bd701, Bd900, Bd1200 and Bd3113), 4 were of the 2n = 20 cytotype (Bd114, Bd115, Bd129 and Bd485), and the remaining 13 were of the 2n = 30 cytotype (Bd146, Bd176, Bd217, Bd220, Bd238, Bd250, Bd260, Bd341, Bd343, d361, Bd409, Bd486 and Bd3107). It should be noted that the populations from the higher interior of the Iberian Peninsula were more commonly diploid (2n = 10) (samples collected in the Provinces of Jaén, Albacete, Cuenca and Guadalajara; see Fig. 1).
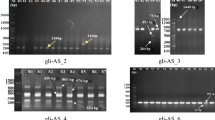
The prolamins of a total 289 plants belonging to all the studied cytotypes were examined by denaturing SDS-PAGE. Twenty-two main bands, ranging from 22.14 to 89.2 kD, were clearly visualized (Fig. 2). The pattern of bands varied between plants belonging to the same population and between populations. The number of bands for the 2n = 10 plants varied from 4 to 8, that of the 2n = 20 plants from 3 to 10 and that of the 2n = 30 plants from 5 to 11. These proteins have similar patterns in the LMW-GS region as Triticum aestivum cv. ‘Chinese Spring’ but lack clear protein bands in the HMW-GS region. The number of total proteins by population varied from 4 (Bd160, Bd162, Bd1200 with 2n = 10) to 16 (Bd409, 2n = 30) or 17 (Bd220, 2n = 30). Some proteins, such as LMW-GS 1, 9A, 10 or 10A, were present in very few populations. Others, such as LMW-GS 11 or 12, were absent in some populations. In general, each population showed a predominant pattern of glutenin subunits and each population was characterized by a mixture of a small number of different patterns. LMW-GS 11, 12, 17 and 18A were the most common among the material examined, and LMW-GS 1, 2, 6, 9, 9A, 13, 14 and 15 the least common. LMW-GS 10 was only present in the diploid accession Bd169. LMW-GS 17 and 17A and 18 and 18A showed a pattern of presence/absence mutually excluding.
The glutenins of B. distachyon. a Patterns of LMW-GS in different plants of three wild populations with 2n = 10. Marks in green indicate bands that are present in forms with 2n = 10 and 2n = 30, but absent in 2n = 20; b variation of storage proteins from total caryopses of different plants of Bd129 (2n = 20). Marks in red indicate bands that are present in forms with 2n = 20 and 2n = 30, but absent in 2n = 10. c Variation of storage proteins from total caryopses of 14 plants of Bd250 (2n = 30). CS glutenins of T. aestivum cv. ‘Chinese Spring’, M molecular weight markers
The 22 distinguishing proteins constitute a set of markers that could be used in the analysis of the diversity within and between populations. Table 2 summarizes the LMW glutenin diversities conducted by Nei’s index, taking into account the set of bands detected in the 12 plants analyzed in each population. The populations have been ordered according their cytotypes. The final rows of each group present the average Nei index for the set of protein subunits in the different cytotypic groups. The last column records the mean heterogeneity index for each population, which varied from 0.01 to 0.44. Interestingly, the 2n = 10 forms showed the lowest Nei indices (from 0.01 in Bd701 and Bd900 to 0.09 in Bd160 and Bd169, with average of 0.05), whereas those with largest number of chromosomes were the most heterogeneous having higher Nei indices (average of 0.12 and 0.17 for the cytotypic groups of 2n = 20 and 2n = 30, respectively.
Of the 22 total LMW-GS found in the 23 natural populations analyzed, only subunit 10 was specifically found at the 2n = 10 level (Bd169). A maximum of 21 bands was recorded for the 2n = 30 forms. Of the LMW-GS, 1, 9A and 15 were present in the 2n = 10 forms and absent in the 2n = 20 forms, 2, 3, 10A, 13, 17A and 18 were present in the 2n = 20 forms but absent in the 2n = 10 forms, and 4, 5, 6, 7, 8, 9, 11, 12, 14, 16, 17 and 18A were common to all forms. The confluence in the 2n = 30 forms of different proteins found in the 2n = 10 and 2n = 20 forms sustains the hypothesis of the origin of the 2n = 30 forms in the addition of the chromosomal complements of the 2n = 10 and 2n = 20 forms.
Figure 3 shows an example of the results obtained with population Bd146 (2n = 30) after FISH with the rDNA probes pTa71 and pTa794, followed by reprobing the same chromosome preparations with genomic DNA from Bd3113 (2n = 10) and Bd129 (2n = 20). FISH with pTa71 and pTa794 showed four signals each on eight different chromosomes. The pTa71 signals, two stronger and two weaker, were located at the distal region of two pairs of small chromosomes. The stronger 5S rDNA locus was found in a proximal region of a medium metacentric chromosome, while the fainter one was visualized in the distal region of a small pair of chromosomes (Fig. 3a). After reprobing with 2n = 10 cytotype genomic DNA, two groups of chromosomes became distinguishable via their fluorescence patterns. Twenty small chromosomes showed fluorescence signals exclusively around the centromeres, and 10 chromosomes of different sizes showed a dispersed pattern of signals along the length of their arms (Fig. 3b). Reprobing metaphase chromosomes of 2n = 30 plants with 2n = 20 cytotype genomic DNA revealed signals exclusively localized at the centromeres of 20 small chromosomes. The merged images clearly distinguished two groups of differentiated chromosomes, 10 larger ones of different size and 20 smaller ones of similar size. Comparison of the results of the sequential hybridization experiments showed that the faint pTa71 and the proximal pTa794 signals were located on chromosomes belong to the group of 10 chromosomes, while the stronger signals observed with pTa71 and the distal pTa794 mapped to chromosomes of the 20 chromosome group (compare Figs. 3a and 3b).
Confirmation that the 2n = 30 B. distachyon cytotype is of alloploid origin. a FISH of the rDNA probes pTa71 (four red signals) and 5S rDNA pTa794 (four green signals) to metaphase of Bd146 (2n = 30). b The same metaphase after reprobing with genomic DNA from Bd3113 (2n = 10) (green) and Bd129 (2n = 20) (red signals). Scale bar = 10 µm
The data matrix on the presence/absence of the 22 LMW-GS in each population was used to compute the output matrix of Dice’s similarity coefficient for interval measure among populations of all cytotypic groups. The coefficients were used to produce a cluster using the unweighted pair-group method (UPGMA). The data derived from the analysis of similarity were used to construct dendrograms. The dendrogram of Fig. 4 is based on Dice’s coefficient (other functions were also essayed, such as Jaccard, giving a similar dendrogram). The following should be noted: (a) there is considerable ramification, reflecting the great variation both within and between populations for the endosperm proteins; (b) two well-defined groups separating the 2n = 10 forms from the 2n = 20 plus 2n = 30 forms can be clearly appreciated; (c) the branch that groups the populations with more than 10 chromosomes shows two clear subgroups separating the populations with 2n = 20 (upper part of the dendrogram) and 2n = 30 forms (middle of the dendrogram) (Fig. 5).
Correspondence analysis displaying the distribution of 23 wild populations of B. distachyon. Six climatic and eco-geographical variables have been contemplated. The populations distribute themselves in two sub-groups that delimit the forms of 2n = 10 from the others (2 n = 20 and 2n = 30). The forms with 2n = 20 have been marked with asterisk
Discussion
The oldest classification of wheat seed proteins is based on their different dilution properties (Osborne 1924) and involves four main fractions: albumins (soluble in water), globulins (soluble in saline solutions), gliadins (soluble in diluted alcohol) and glutenins (soluble in acid solutions or alkalis). Even though the four types show clear biochemical differences, their solubility in different solvents is not absolutely discriminatory. The major protein groups of the endosperm of wheat are the glutenins and gliadins, which together are known as prolamins owing to their high content of proline and glutamine. They make up about 80% of the total protein of the grain, are synthesized in the endoplasmic reticulum in the final stages of maturation and are deposited as the main nitrogen reserve in the cytoplasm in the form of “protein bodies” for mobilization and use during germination (Shewry and Mifflin 1985).
A recent study on major seed storage proteins in a diploid accession of this species reports the main protein component to involve the 12S and 7S globulin types (Laudencia-Chingcuanco and Vensel 2008). A subset of the major seed proteins isolated from three hexaploid accessions was been also identified as globulins. Larré et al. (2010) recently studied the protein composition of caryopses of a 2n = 10 accession of Brachypodium by separating the proteins on the basis of their solubility, combining this with proteomic analyses. These authors detected globulins and prolamins, but no albumins. Globulins were represented mainly by the 11S type, and their solubility properties corresponded to the glutelin found in rice. Wang et al. (2010) found abundant wheat LMW-GS-like proteins and a few HMW-GS with low levels of expression in three 2n = 10 and 10 accessions of B. distachyon with 2n = 30 analyzed by SDS-PAGE and other techniques. In total, more than 20 different bands in the LMW-GS region and 15 albumin proteins were identified in these materials.
The present study followed a specific method to extract the glutelins, which were then separated by SDS-PAGE. The result was the identification of at least 22 distinguishable bands corresponding to the wheat LMW-GS-like proteins. These showed a high degree of variability within and between the 2n = 10, 2n = 20 and 2n = 30 accessions investigated. These proteins must be considered wheat LMW-GS-like due to the method of extraction that released them and their correspondence with the proteins characterized by Wang et al. (2010).
Protein variability in the populations of B. distachyon has been poorly documented to date. The present work, however, reveals the great variation of endosperm proteins shown across the species. A population is considered polymorphic when there is variation in at least one band and/or pattern in at least one individual in a population (Asíns and Carbonell 1986; Soler et al. 1993). In agreement with this criterion, all the natural populations investigated can be classed as polymorphic for the endosperm proteins studied.
The 2n = 30 forms showed the greatest diversity, followed by the 2n = 20 and finally the 2n = 10 forms. According to Nevo (1978), the variability found in autogamous species could be due to their location. Thus, those populations with wide distributions ought to be relatively more polymorphic and heterogeneous than those with more restricted distributions. The natural populations of an autogamous species tend to be constituted by homozygous genotypes. The LMW-GS-like proteins observed in the natural populations of B. distachyon indicate the existence of a mixture of plants within each population, although there was always a predominant pattern. The different patterns might represent distinct homozygous genotypes, which might be maintained from one generation to the next as pure lines by self-pollination. Certainly, the natural populations of B. distachyon showed more variation than expected with regard to their autogamous breeding system. In agreement with Stebbins (1975), it is to be expected that when an autogamous species extends towards new habitats that it should establish a greater number of different lines than would a population that for generations has remained in the same habitat. This might be the circumstances of the populations of B. distachyon in the Iberian Peninsula, near the limit of the species’ distribution. It is also possible, however, that the difference in variability is the consequence of different degrees of allogamy influenced by environmental factors (Sanders and Hamrick 1980; Schoen and Clegg 1985; Golenberg 1988; Monte et al. 1999; Nieto et al. 2000, 2003).
Vogel et al. (2009) characterized the natural diversity in Turkish populations of B. distachyon using simple sequence repeat (SSR) markers and demonstrated that under growth chamber and greenhouse conditions the flowers of 2n = 10 plants rarely open. Only under certain environmental conditions (warm and humid in full sun) were open flowers observed on 2n = 10 plants growing outside greenhouse. The same authors monitored the rate of pollen flow using potential transgenic pollinators and demonstrated that the rate of outcrossing under laboratory conditions is exceedingly small. The same authors reported that ‘polyploid’ forms show exerted anthers, which could imply the existence of allogamy in these cases. This, and the possession of duplicated genomes, could explain why the 2n = 30 plants present more diversity.
To analyze the influence of eco-geographic and climatic factors in the distribution of the studied populations, analysis of correspondence was performed using a two-way contingency matrix that included the number of chromosomes and the data of six geographic and climatic variables corresponding to the sites of natural populations. The results indicate the main factors that influence the three first axes of correspondence (96.58% of the total variance) to be altitude, latitude, summer precipitation, the index of Mediterraneity, the thermicity index and the annual mean temperature.
An analysis was then made of the possible correlation between the mentioned eco-geographic and climatic factors and the diversity shown by the endosperm proteins in the populations, represented by the Nei index (Table 3). The diversity correlated positively with the number of chromosomes (2n), annual mean temperature, index of thermicity and index of Mediterraneity. In contrast, diversity correlated negatively with altitude, latitude and index of summer precipitation. The number of chromosomes correlated negatively with altitude, latitude and summer precipitation and positively with all the other climatic indices. These results are coherent with the fact that diploid forms were more common in the populations collected from the interior and at higher altitude, where the climate is more extreme.
The present study highlights the habitat preferences and degree of heterogeneity shown by the forms of B. distachyon with different numbers of chromosomes as determined by DAPI staining, FISH and GISH. The two small pairs of chromosomes of the 2n = 10 cytotype bear a 45S locus in the distal region of the short arm and a major 5S rDNA locus in the proximal region of the long arm. These sequences are located in the distal regions of two different pairs of chromosomes in the 2n = 20 cytotype. The chromosomes of the 2n = 20 form are considerably smaller than those of the 2n = 10 form. The 2n = 30 cytotype shows the same characteristics of chromosome size and positioning as the 45S and 5S rDNA sequences of both the 2n = 10 and 2n = 20 forms, suggesting it to be alloploid in origin as proposed by Hasterok et al. (2004).
These differences seem to corroborate the proposal made by Hasterok et al. (2004) that the forms of Brachypodium with different chromosome number be separated into different taxonomic units. This separation has also been suggested on the basis of C-values and the different size of the sets of chromosomes observed by FISH. Hasterok et al. (2004) used FISH to analyze different accessions of Brachypodium and found the 2n = 20 forms to have small chromosomes more similar to those of B. sylvaticum than to the larger chromosomes found in 2n = 10 accessions of B. distachyon. Moreover, 2n = 30 accessions are reported to have C-values approximately twice as large as those for the 2n = 10 forms (Vogel et al. 2006), and their karyotypes seem to represent the addition of the chromosomes of the 2n = 10 and 2n = 20 forms. All this suggests that the 2n = 30 forms are allotetraploids formed by hybridization between 2n = 10 and 2n = 20 forms, followed by chromosome doubling (Hasterok et al. 2004). Moreover, all three cytotypes with 2n = 10, 20 and 30 chromosomes in this study belong to B. distachyon and have been collected in the Iberian Peninsula. Thus, the alloploid origin proposed by Hasterok et al. (2004) is possible also within B. distachyon with different cytotypes. The smaller number of endosperm protein subunits shared by the 2n = 10 and 2n = 20 forms than between either of the above and the 2n = 30 forms corroborates the possible allopolyploidal origin of the 2n = 30 forms. Some authors have already proposed the classification of the different ploidy forms of B. distachyon as a different species (Hasterok et al. 2004, 2006; Idziak and Hasterok 2008).
References
Asíns MJ, Carbonell EA (1986) A comparative study on variability and phylogeny of Triticum species. 1. Intraspecific variability. Theor Appl Genet 72:551–558
Catalan P, Olmstead RG (2000) Phylogenetic reconstruction of the genus Brachypodium. Beauv. (Poaceae) from combined sequences of chloroplast gene and nuclear ITS. Plant Syst Evol 220:1–19
Catalan P, Kellogg EA, Olmstead RG (1997) Phylogeny of Poaceae subfamily Pooideae based on chloroplast ndhF gene sequences. Mol Phylogen Evol 8:150–166
Cuadrado A, Acevedo R, Moreno Diaz De La Espina S, Jouve N, De La Torre C (2004) Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J Exp Bot 55:847–854
Doust A (2007) Architectural evolution and its implications for domestication in grasses. Ann Bot 100:941–950
Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM (2001) Brachypodium distachyon a new model system for functional genomics in grasses. Plant Physiol 127:1539–1555
Filiz E, Ozdemir BS, Budak F, Vogel JP, Tuna M, Budak H (2009) Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52(10):876–890
Garvin DF (2007) Brachypodium: a new monocot model plant system emerges. J Sci Food Agric 87:1177–1179
Garvin DF, Gu Y, Hasterok R, Hazen S, Jenkins G, Mockler T, Mur LJ, Vogel JP (2008) Development of genetic and genomic research resources for Brachypodium distachyon, a new model for grass crop research. Crop Sci 48:S69–S84
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat that contain 5S rDNA genes. Nucleic Acids Res 8:4851–4865
Golenberg EM (1988) Outcrossing rates and their relationship to phenology in Triticum dicoccoides. Theor Appl Genet 75:937–944
Hasterok R, Draper J, Jenkins G (2004) Laying the cytotaxonomic foundations of a new model grass Brachypodium distachyon (L.) Beauv. Chromosom Res 12:397–403
Hasterok R, Marasek A, Donnison IS, Armsted I, Thomas A, King IP, Wolny E, Idziak D, Draper J, Jenkins G (2006) Alignement of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridisation. Genetics 173:349–362
Hawkes JG (1980) Crop genetic resources. Field collection manual for seed crops, roots and tuber crops, tree fruit crops and related wild species. IBPGR/EUCARPIA, Rome
Higgins JA, Bailey PC, Laurie DA (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5(4):e10065. doi:10.1371/journal.pone.0010065
Huo N, Lazo G, Vogel J, You F, Ma Y, Hayden DM, Coleman-Derr D, Hill T, Dvorak J, Anderson OD, Luo M, Gu YQ (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC and sequences. Funct Integr Genomics 8:135–147
Huo N, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, Dvorak J, Anderson OD, Luo MC, Gu YQ (2009) Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol Biol 70:47–61
Idziak D, Hasterok R (2008) Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 51:387–391
Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125:1198–1205
Larré C, Penninck S, Bouchet B, Lollier V, Tranquet O, Denery-Papini S, Guillon F, Rogniaux H (2010) Brachypodium distachyon grain: identification and subcellular localization of storage proteins. J Exp Bot 61(6):1771–1783. doi:10.1093/jxb/erq050
Laudencia-Chingcuanco DL, Vensel WH (2008) Globulins are the main seed storage proteins in Brachypodium distachyon. Theor App Genet 117:555–563
Monte JV, Casanova C, Soler C (1999) Genetic variation in Spanish populations of the genus Aegilops revealed by RAPDs. Agronomie 9:419–427
Nevo E (1978) Genetic variation in natural populations: pattern and theory. Theor Populat Biol 13:121–177
Nieto RM, Casanova C, Soler C (2000) Analysis of the genetic diversity of wild Spanish populations of the species Elymus caninus (L.) Linnaeus and Elymus hispanicus (Poiss) Talavera by PCR-based markers and endosperm. Agronomie 21:893–905
Nieto RM, Soler C, García P (2003) Genetic diversity in wild Spanish populations of Thinopyrum junceum and Thinopyrum junceiforme using endosperm proteins and PCR-based markers. Hereditas 139:18–27
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177
Osborne TB (1924) The vegetable proteins, 2nd ed edn. Longmans Green, London
Ozdemir BS, Hernandez P, Filiz E, Budak H (2008) Brachypodium genomics. Int J Plant Genomics 536104. doi:10.1155/2008/536104
Rivas-Martínez S, Lousa M, Díaz TE, Costa J (1990) La vegetación del Sur de Portugal (Sado, Alentejo y Algarve). Itin Geobot 3:5–126
Robertson IH (1981) Chromosome numbers in Brachypodium (Poaceae). Plant Syst Evol 188:125–138
Rolf JF (1994) NTSYS-pc. Numerical taxonomy and multivariate analysis system. Pc versión. 2.11. Exeter Software Applied Biostatistics Inc, New York
Sanders TB, Hamrick JL (1980) Variation in the breeding system of Elymus Canadensis. Evolution 34:117–122
Schoen DJ, Clegg MT (1985) The influence of flower colour on outcrossing rate and male reproductive success in Ipomoea purpurea. Evolution 39:1242–1249
Shewry PR, Mifflin BJ (1985) Seed storage proteins of economically important cereals. Adv Cereal Sci Technol 7:1
Shewry PR, Tathan AS (1995) Seed storage proteins: structure and biosynthesis. Plant Cell 7:945–956
Shi Y, Draper J, Stace C (1993) Ribosomal DNA variation and its phylogenetic implication in the genus Brachypodium (Poaceae). Plant Syst Evol 188:125–138
Singh NK, Shepherd KW, Cornish GB (1991) A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J Cereal Sci 14:203–208
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Soler C, Galindo C, Garcia P, Gonzalez-Castaño S, Casanova C (1993) Isozyme variation in Spanish natural populations of Elytrigia pungens (pers.) and Elytrigia repens (L.) Newski. Heredity 71:51–58
Soler C, Casanova C, Rojo A (2004) Desarrollo de cubiertas vegetales a partir de gramíneas seleccionadas, para su explotación en tierras de olivar. Actas Hortic 41:97–100
Sorensen T (1948) A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons K. Dan. Vidensk. Selsk. Biol Shar 5:1–34
Stebbins GL (1975) The role of polyploid complexes in the evolution of North American grasslands. Taxon 24:91–106
Thornthwaite CW (1948) An approach toward a rational classification of climate. Geograph Rev 38(1):55–94
Vain P, Worland B, Thole V, Mckenzie N, Alves SC, Opanowicz M, Fish L, Bevan M, Snape JW (2008) Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd 21) for T-DNA insertional mutagenesis. Plant Biotech J 6:236–245
Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21–3. Plant Cell Rep 27:471–478
Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D (2006) EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor Appl Genet 113(2):186–195
Vogel JP, Tuna 1M, Budak H, Huo N, Gu YQ, Steinwand MA (2009) Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol 9:88. doi:10.1186/1471-2229-9-88
Wang K, Han X, Dong K, Gao L, Li H, Ma W, Yan Y (2010) Characterization of seed proteome in Brachypodium distachyon. J Cereal Sci 52, doi:10.1016/j.jcs.2010.05.001
Acknowledgments
This study was supported by grants from the Spanish Ministry of Science and Innovation (AGL 2009-10373). The authors thank Adrian Burton for linguistic assistance.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hammami, R., Jouve, N., Cuadrado, A. et al. Prolamin storage proteins and alloploidy in wild populations of the small grass Brachypodium distachyon (L.) P. Beauv.. Plant Syst Evol 297, 99 (2011). https://doi.org/10.1007/s00606-011-0502-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-011-0502-7