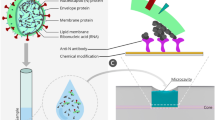
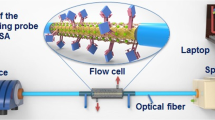
Abstract
The label-free detection of SARS-CoV-2 spike protein is demonstrated by using slightly tapered no-core fiber (ST-NCF) functionalized with ACE2. In the fabricated sensor head, abrupt changes in the mode-field diameter at the interfaces between single-mode fiber and no-core fiber excite multi-guided modes and facilitate multi-mode interference (MMI). Its slightly tapered region causes the MMI to be more sensitive to the refractive index (RI) modulation of the surrounding medium. The transmission minimum of the MMI spectrum was selected as a sensor indicator. The sensor surface was functionalized with ACE2 bioreceptors through the pretreatment process. The ACE2-immobilized ST-NCF sensor head was exposed to the samples of SARS-CoV-2 spike protein with concentrations ranging from 1 to 104 ng/mL. With increasing sample concentration, we observed that the indicator dip moved towards a longer wavelength region. The observed spectral shifts are attributed to localized RI modulations at the sensor surface, which are induced by selective bioaffinity binding between ACE2 and SARS-CoV-2 spike protein. Also, we confirmed the capability of the sensor head as an effective and simple optical probe for detecting antigen protein samples by applying saliva solution used as a measurement buffer. Moreover, we compared its detection sensitivity to SARS-CoV-2 and MERS-CoV spike protein to examine its cross-reactivity. In particular, we proved the reproducibility of the bioassay protocol adopted here by employing the ST-NCF sensor head reconstructed with ACE2. Our ST-NCF transducer is expected to be beneficially utilized as a low-cost and portable biosensing platform for the rapid detection of SARS-CoV-2 spike protein.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic information of the new coronavirus of severe acute respiratory syndrome (SARS) was released to the world in January 2020. The World Health Organization termed the discovered novel virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2,3]. Since the structure of the receptor-binding gene of SARS-CoV-2 was very similar to that of SARS-CoV, it was speculated that SARS-CoV-2 would use the same receptor as SARS-CoV when infiltrating cells, and in later studies, it was proved that SARS-CoV-2 also uses angiotensin-converting enzyme 2 (ACE2) as the receptor [4,5,6]. In the case of other viruses, middle east respiratory syndrome coronavirus (MERS-CoV) and alpha coronavirus (Alpha-CoV) use dipeptidyl-peptidase 4 (DPP4) and aminopeptidase N (APN) as their receptors, respectively [7, 8]. Human coronavirus NL63 (HCoV-NL63) can also utilize ACE2 but has a lower binding affinity compared to SARS-CoV [9]. Coronaviruses are spherical structures with a diameter of 80 to 125 nm and have a viral envelope derived from the plasma membrane of the host cell. The viral envelope consists of three viral structural proteins of spike protein (S), membrane protein (M), and envelope protein (E), and it contains a nucleocapsid protein in the membrane. In particular, the spike protein of coronaviruses has been known to be essential for the virus to penetrate the cell by interacting with the receptor of the host cell [10, 11]. Surprisingly, some studies have reported that SARS-CoV-2 spike protein has a greater binding affinity for ACE2 than SARS-CoV spike protein. SARS-CoV-2 mutants may reinforce the interaction between SARS-CoV-2 spike protein and ACE2. Real-time polymerase chain reaction (RT-PCR) is a gold-standard method because of its high selectivity and sensitivity for the detection of COVID-19 infection [12]. Due to the false-negative results of RT-PCR, there has been an increasing interest in the biosensor as an alternative and first test [13,14,15,16,17,18,19,20]. Recently, it was found that SARS-CoV-2 spike protein could bind with human ACE2 protein in a functional ELISA assay. Qiu et al. demonstrated a dual-functional localized surface plasmon resonance (LSPR) biosensor in combination with a plasmonic photothermal (PPT) effect, showing sensitivity to SARS-CoV-2 viral nucleic acid with a limit of detection (LOD) of 0.22 pM [15]. For rapid diagnosis of SARS-CoV-2 spike protein, Seo et al. reported graphene-based field-effect transistor biosensors functionalized with SARS-CoV-2 spike antibody and achieved an LOD of 1.6 × 101 pfu/mL in culture medium and an LOD of 2.42 × 102 copies/mL in clinical samples, respectively [16]. More recently, Raziq et al. proposed an electrochemical biosensor integrated with molecularly imprinted polymers capable of detection of SARS-CoV-2 nucleoprotein [19], which showed a linear response in detecting SARS-CoV-2 protein with an LOD of 15 fM.
Specifically, in the field of label-free biosensing dealing with various types of biomaterials including viruses, optical fiber sensors have been studied steadily and attracted much attention as important platforms to detect bioelements due to their immunity to electromagnetic interference, electrical passivity, chemical inertness, multiplexing capability, and fast response [21, 22]. The optical biosensing platform relies on the measurement of changes in the localized refractive index (RI) of the surface due to the binding between biorecognition elements and bioanalytes. Optical transducers based on this principle include long-period fiber gratings (LPFGs) [23,24,25], fiber Bragg gratings (FBGs) [26], surface plasmon resonance (SPR)-based fiber-optic probe [27], U-bent fiber [28], fluorescence-based fiber-optic probe [29], and tapered optical fiber (TOF) [30, 31], which aim to improve the sensitivity of the bioreaction. For example, Wang et al. demonstrated an SPR biosensor based on graphene oxide/silver-coated polymer cladding silica fiber, whose surface was immobilized with anti-human immunoglobulin G (IgG), for the detection of human IgG [27]. Wen et al. reported a U-shaped optical fiber biosensor coated with miRNA-133a for the detection of microRNA, Mahlavu specimen [28]. Mustapha Kamil et al. demonstrated a tapered single-mode fiber (SMF) sensor immobilized with dengue virus E protein antibodies for the detection of dengue virus [30]. Chen et al. developed a tapered no-core fiber (NCF) coupler sensor functionalized with pig IgG antibodies for the detection of S. aureus [31]. This TOF sensor head with a tapered diameter of 10.4 μm is highly sensitive to the surrounding medium RI. However, at the same time, it is considered that the sensor head is very difficult to handle because it is so vulnerable to transverse stress that can be exerted by slight bending or twisting and weak tensile strain. As such, although several studies related to the TOF sensor have been steadily conducted owing to the advantage of its high sensitivity in diverse fields measuring chemical or mechanical quantities including the biosensing area, there is still a crucial issue of the vulnerability of the tapered section, which deteriorates the sustainability and stability of the fabricated sensor.
Here we present a slightly tapered NCF (ST-NCF) whose either end is concatenated with SMF as an optical fiber transducer for the label-free detection of SARS-CoV-2 spike protein. To obtain a fiber sensor head more robust than other TOF sensors, the NCF segment in the fabricated transducer was tapered at a cladding diameter of ~ 100 μm using a commercial standard fiber fusion splicer in a simple and reproducible way. Abrupt changes in the mode-field diameter at the interfaces between SMF and NCF excite multiple propagation modes, and multi-guided modes create multi-mode interference (MMI) when they meet at the lead-out SMF. The surface of the sensor head was functionalized with human ACE2 bioreceptors, which can specifically bind to SARS-CoV-2 spike protein. This is the first study to diagnose SARS-CoV-2 spike protein utilizing the ST-NCF conjugated with human ACE2 protein. This study has the significance that the fabricated ST-NCF is not only relatively robust to transverse stress but also sensitive enough to target viral envelope proteins after proper biofunctionalization. In the MMI spectrum of the functionalized sensor head, we selected the transmission minimum as a sensor indicator dip reflecting the bioaffinity interaction between bioreceptors and target analytes. To investigate the detection capability of SARS-CoV-2 spike protein, the wavelength shift of the sensor indicator was monitored for spike protein sample solutions with different concentrations in PBS and saliva buffer. In particular, to determine whether the sensor response obtained by detection of SARS-CoV-2 spike protein is attributed to its specific interaction with ACE2 bioreceptors, we checked the cross-reactivity of our sensor head by measuring its response to MERS-CoV spike protein as well as SARS-CoV-2 spike protein. Moreover, we proved the reproducibility of the bioassay protocol adopted here by assessing the performance of the sensor head reconstructed by bioconjugating the used ST-NCF with ACE2 again.
Experimental
Reagents and materials
Sulfuric acid (H2SO4), hydrogen peroxide (H2O2), and ethanol were purchased from SAMCHUN chemicals (Seoul, Korea). (3-aminopropyl) triethoxysilane (APTES), glutaraldehyde (GA), and phosphate buffered saline (10 mM PBS, pH 7.4) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Distilled water, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), and N-hydroxysulfosuccinimide (Sulfo-NHS) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Human ACE2 protein (10,108-H05H) and SARS-CoV-2 spike protein (40,589-V08B1) were purchased from Sino Biological (Beijing, China). MERS-CoV spike protein (MERSS126-R-10), saliva samples (991–05-P), and bovine serum albumin (BSA, A0100-010) were purchased from Alpha Diagnostic Intl (TX, USA), Lee Biosolutions (MO, USA), and GenDEPOT (Katy, TX, USA), respectively.
Fabrication of ST-NCF sensor head and principle of operation
As the first step for the fabrication of the ST-NCF sensor head, either end of an NCF (FG125LA, Thorlabs) segment was fusion-spliced with SMF. The cladding diameter of the NCF was 125 μm, and the optimal length of the NCF was tailored to be 28 mm to obtain MMI spectral dips whose wavelengths fall within the output wavelength range of a broadband light source used here for biosensing. When light propagating in the input SMF enters the NCF, a series of higher-order modes are excited within the NCF region due to the mode-field mismatch, followed by the interference between the fundamental and high-order modes, which occurs as light travels to the output SMF [32, 33]. The NCF with its either end concatenated with SMF can be regarded as an MMI apparatus. Because the surrounding medium of this MMI apparatus can act as a cladding layer of the NCF, the guided modes within the NCF are easily affected by the RI change of the surrounding medium and thus a significant shift in the wavelength of MMI created here can be induced according to external perturbations in the surrounding medium RI. In the waveguide model based on the far-from-cut-off approximation for NCF [34], which is modified from the weakly guiding approximation applied for the modal analysis of multi-mode fiber [35, 36], the normalized optical frequency is given by
where α, λ, nncf, and nmedium are the radius of NCF, the free-space wavelength, the NCF RI, and the surrounding medium RI, respectively [37]. Thus, a change in V, which can be induced by the presence of mechanical deformation or surface RI fluctuation at the NCF region of the MMI apparatus, is now associated with the increased evanescent-wave portion of propagation modes due to coupling of multiple higher-order modes, which is responsible for sensitivity enhancement. Assuming that the SMF and NCF are ideally aligned so that the input field becomes circularly symmetric, only LP0m mode (i.e., the high-order modes of NCF) can be excited [22, 38]. The output transmittance (T) of the MMI apparatus (i.e., SMF-NCF-SMF structure) can be determined as
where M is the total number of modes of the NCF section, L is the length of the NCF section, c0m is the excitation coefficient of the LP0m mode, and β0m is the propagation constant of the LP0m mode [38].
Furthermore, the incorporation of tapering into NCF-based sensors increases the magnitude of the evanescent field and thus its penetration depth, allowing for more sensitive and accurate measurement of physical or chemical parameters applied to the sensor surface [37]. An optical fiber transducer utilizing TOF is based on evanescent wave sensing, which is one of the transduction mechanisms for detecting biomolecules. As the tapering process increases the evanescent field and exposes it to the surrounding medium, the sensitivity of the TOF sensor can be improved, so when investigating analytes, increased modulation depths in the output signal can be observed with respect to the RI change at the surrounding medium [39]. In our sensor head composed of ST-NCF and SMF (concatenated with either end of the ST-NCF), the ST-NCF segment was fabricated by tapering NCF with a fiber fusion splicer. The tapering method based on a fusion splicer has some advantages of convenient control and good repeatability compared to other methods, such as flame heating [40, 41], chemical etching [42, 43], and CO2 laser illumination [44, 45]. Optimized tapering conditions are described in detail in “Tapering conditions of no-core fiber” of the Supplementary Information. Figure 1a shows a schematic diagram of the geometrical structure of the fabricated ST-NCF sensor head. The tapered waist diameter and the tapered length of the ST-NCF segment are ~ 103 and ~ 340 μm, respectively. Figs. S1a and S1b show a low-vacuum scanning electron microscope (LV-SEM, JSM-6490LV, JEOL) image and an optical microscope image of the fabricated ST-NCF sensor head, respectively. The prepared ST-NCF sensor head will be biofunctionalized with receptors for SARS-CoV-2 spike protein detection through the subsequent surface treatment process. Before the surface modification of the ST-NCF sensor head, the RI response of the sensor indicator, i.e., the transmission minimum near ~ 1516 nm in air, selected in the MMI spectrum of the ST-NCF sensor head, was scrutinized in an RI range of 1.333 to 1.420 at room temperature. As shown in Fig. 1b and c, when the concentration of sucrose in aqueous solution increases from 0 to 50%, that is, when the surrounding medium RI of the ST-NCF sensor head increases, the indicator dip moves towards a longer wavelength region showing a total wavelength shift of ~ 10.84 nm. The inset of Fig. 1b is its zoomed version plotted over a wavelength range from 1510 to 1550 nm. The RI sensitivity of the ST-NCF sensor head was measured as 126 nm/RIU with an adjusted R2 value of 0.9923.
a Schematic diagram of fiber structure of fabricated ST-NCF sensor head. b Spectral variations of indicator dip of fabricated (bare) ST-NCF sensor head, measured for external RI changes of 1.333 to 1.420 at room temperature. The inset shows the zoomed spectra plotted over a wavelength range from 1510 to 1550 nm. c RI-induced wavelength shifts of sensor indicator. A solid line represents the linear fit of measured data (i.e., indicator wavelength)
Surface functionalization of ST-NCF sensor head
To introduce biosensitivity onto the fabricated ST-NCF sensor head, we need a pretreatment process to functionalize the sensor head surface with bioreceptors, which can specifically bind to SARS-CoV-2 spike protein. Figure 2a shows a schematic diagram of the surface functionalization process of the ST-NCF sensor head. The functionalization process is composed of the following steps: (1) Piranha cleaning and washing, (2) APTES silanization, (3) GA treatment, (4) EDC/Sulfo-NHS activation, (5) ACE2 immobilization, (6) removal of unbound ACE2, and (7) BSA blocking of non-specific binding sites. Through the above sequential functionalization procedure, the ST-NCF sensor head functionalized with ACE2 was prepared as a biosensing immunosensor for the detection of target analytes. Figure 2b represents the transmission spectrum of the ST-NCF sensor head, measured in air before and after the immobilization of ACE2 bioreceptors on its surface, displayed with black and green solid lines, respectively. The inset of Fig. 2b shows the magnified transmission spectrum near the transmission minimum within a wavelength range from 1460 to 1480 nm. It was observed that the transmission minimum near 1468 nm of the bare ST-NCF sensor head shifted by ~ 1.38 nm towards a longer wavelength region after the completion of the functionalization process owing to the effect of the surface treatment. Although the degree of the wavelength shift was different, this wavelength shift of the transmission dip was also observed after individual functionalization processes, including the APTES silanization, the GA treatment, the EDC/Sulfo-NHS activation, and the ACE2 immobilization processes.
a Schematic diagram of surface functionalization process of fabricated ST-NCF sensor head for detection of SARS-CoV-2 spike protein: APTES silanization, GA treatment (followed by EDC/Sulfo-NHS activation), immobilization of ACE2 bioreceptors, BSA blocking, and bioreaction with SARS-CoV-2 spike protein (in sequence). b Transmission spectrum of ST-NCF sensor head measured in air before and after immobilization of ACE2 bioreceptors on its surface, displayed with black and green solid lines, respectively. The inset shows the magnified transmission spectrum near the transmission minimum within a wavelength range from 1460 to 1480 nm
Measurement setup for detection of target analytes
Figure 3 shows the experimental setup for the detection of SARS-CoV-2 spike protein using the ACE2-immobilized ST-NCF sensor head. A broadband light source (Fiberlabs FL7701) and an optical spectrum analyzer (Yokogawa AQ6370C) were utilized for the measurement of the perturbation-induced variations in the transmission spectrum of the biofunctionalized sensor head. Because the broadband output of the light source is conveyed to the optical spectrum analyzer via the optical fiber waveguide of the sensor head, we can monitor the spectral variations, especially wavelength shifts, of the indicator dip in the transmission spectrum, induced by bioreaction occurring at the surface of the sensor head. The spectrum analyzer was remote-controlled, and its spectral data were acquired in real-time by a laptop using LabVIEW. As a final step to quantitatively assess the binding reaction between bioreceptors and analytes, we located the ACE2-conjugated ST-NCF sensor head in the glass container and attached its either end to fixed stages so that it remained straight during the spectral measurement. We prepared the solution samples of SARS-CoV-2 spike protein with different concentrations of 1, 101, 102, 103, and 104 ng/mL in turn, by sequentially diluting the stock solution using PBS (10 mM, pH 7.4). Then, we applied the solution samples to the ST-NCF sensor head immobilized with ACE2 of 250 μg/mL in concentration. Because the minimum volume to completely cover the sensor head surface was 100 μL, a SARS-CoV-2 spike protein solution of 100 μL was used for every sample concentration. In the case of the detection of SARS-CoV-2 spike protein in human saliva, saliva used as a measurement buffer was diluted by 1/103 with PBS. Two ST-NCF sensor heads manufactured under the same conditions, including tapering conditions (specifically, fusion splicer conditions) and surface functionalization conditions, were adopted respectively for PBS and saliva experiments. The application of all solution samples was carried out only by pipetting without moving the ST-NCF sensor head until the entire surface of the sensor head was covered with the solution samples. All detection-related procedures described in the next section were performed at room temperature (26 °C).
Results and discussions
Detection of SARS-CoV-2 spike protein in PBS solution
The solution samples of SARS-CoV-2 spike protein were prepared from a high concentration (104 ng/mL) to a low concentration (1 ng/mL) by dilution with PBS. On the contrary, the prepared samples of SARS-CoV-2 spike protein were detected from a low concentration (1 ng/mL) to a high concentration (104 ng/mL) in case of measurement using our ACE2-functionalized ST-NCF sensor head. For every sample concentration, we injected the sample of SARS-CoV-2 spike protein into the glass container, within which the sensor head was located, so that the entire tapered section of the sensor head surface was covered with the sample and then monitored the spectral variation of the indicator dip (i.e., the transmission minimum). After every real-time measurement of the indicator wavelength for ~ 5 min at a specific sample concentration of SARS-CoV-2 spike protein, we vacated the container by taking the solution samples away and rinsed the sensor surface with PBS to remove unbound SARS-CoV-2 spike protein for the following measurement of SARS-CoV-2 spike protein with different concentrations. Before we moved on to the next measurement for the different sample concentration, we reinjected fresh PBS into the container and recorded the post-bioreaction transmission spectrum of the sensor head that had undergone the binding reaction between ACE2 and SARS-CoV-2 spike protein. We iterated this two-step measurement procedure while changing (i.e., increasing) the sample concentration. The post-bioreaction transmission spectrum is slightly different from the transmission spectrum measured when the sensor head is immersed in the sample solution before the PBS washing, which is attributed to the RI difference between the sample solution and PBS solution.
Figure 4a shows the real-time transient wavelength variations of the indicator dip when the sample concentration of SARS-CoV-2 spike protein increases from 1 to 104 ng/mL including the case without application of the solution sample, indicated by 0 ng/mL. Since the optical spectrum analyzer used here takes 7 ~ 8 s to scan the transmission spectrum of the sensor head once, the acquisition interval of the wavelength data in monitoring the indicator wavelength was set to 16 s, about twice the scanning time of the optical spectrum analyzer. It is observed from the figure that the indicator wavelength gradually increases with the sample concentration without significant fluctuations in the wavelength at a fixed sample concentration (in a time scale less than 320 s). From this step-wise transient response, it is confirmed that a noticeable change in the indicator wavelength occurs within seconds after every injection of the solution samples of different concentrations. Moreover, subsequent deviations from the saturated wavelength after the sample injection are not apparent for most sample concentrations, which indicates that our ACE2-functionalized ST-NCF sensor head has rapid and stable responses to SARS-CoV-2 spike protein. In the case of the highest sample concentration (104 ng/mL), the indicator wavelength slightly increases over time unlike the cases of the other sample concentrations, but this transient variation is also stabilized after 150 s. As a result, the single detection time of the proposed sensor can be estimated within 3 min considering the longest stabilization time and including some temporal margins. Figure 4b shows perturbation-induced variations in the transmission spectrum of our sensor head, measured after the real-time observation for every sample concentration with PBS solution used as a measurement buffer. The inset of Fig. 4b shows the magnified transmission spectrum displayed in a wavelength range from 1474 to 1482 nm. As seen in the figure, the indicator dip redshifts from 1475.88 to 1478.26 nm with increasing concentration of SARS-CoV-2 spike protein. With the increase of the sample concentration, the transmittance of the indicator dip also changes from − 37.9 to − 45.85 dB, but its decreasing behavior is not monotonous. In our sensor head, the wavelength shift, rather than the transmittance variation, showed a more consistent and monotonous dependence on the sample concentration. This can also be supported in the following measurement results (i.e., Fig. 5a and b) obtained using the saliva solution. Thus, we focused on the analysis of perturbation-induced variations of the indicator wavelength, instead of the transmission level of the indicator dip.
a Real-time transient wavelength variations of indicator dip for sample concentration of SARS-CoV-2 spike protein increasing from 0 to 104 ng/mL. b Perturbation-induced variations in transmission spectrum of our sensor head, measured after real-time observation for every sample concentration with PBS solution used as measurement buffer. The inset shows the magnified transmission spectrum displayed in a wavelength range from 1474 to 1482 nm. c Indicator wavelength displacement (Δλ) of ACE2-functionalized ST-NCF sensor head and bare ST-NCF sensor head for each applied sample concentration, indicated by navy circles and black triangles, respectively. Navy and black solid lines represent the nonlinear regression results of measured Δλ data. For both sensor heads, the reference wavelength was set as the average of five indicator wavelength values, consecutively measured in PBS without application of the sample solution (i.e., 0 ng/mL), indicated by a black solid line in (b)
a Bioaffinity binding-induced changes in transmission spectrum of ACE2-immobilized ST-NCF sensor head, measured in saliva solution after pre-reaction with SARS-CoV-2 spike protein for every sample concentration. The inset shows the zoomed transmission spectrum observed over a wavelength range from 1510 to 1518 nm. b Indicator wavelength displacement (Δλ) for each applied sample concentration, which was evaluated from measured spectra shown in (a). The reference wavelength was also determined in the same way as used in the case of Fig. 4c. c Cross-reactivity investigation: Indicator wavelength shifts (Δλ’s) induced by reaction between MERS-CoV spike protein and ACE2, which were measured using PBS (blue squares) and saliva (red squares) solution as measurement buffer, respectively. For comparison, Δλ’s caused by the SARS-CoV-2 spike protein binding, measured using PBS and saliva solution, were also displayed with blue and red circles, respectively. d Reusability and reproducibility evaluation: Indicator wavelength displacement according to sample concentration, which is obtained by combining three individual Δλ measurement results given by M-1, M-2, and M-3 in Fig. S3. All three measurements were done with one ST-NCF sensor head, and the whole surface treatment process of the washed sensor head was iterated for every measurement, as shown in the inset. All solid and dashed lines in (b), (c), and (d) are the nonlinear regression results of measured Δλ data
Figure 4c shows the wavelength displacement Δλ of the indicator dip for each applied sample concentration when the detection of SARS-CoV-2 spike protein is carried out using the functionalized ST-NCF sensor head and the bare ST-NCF sensor head, indicated by navy circles and black triangles, respectively. In the evaluation of the wavelength displacement for both sensor heads, the reference wavelength was set as the average of five indicator wavelength values, consecutively measured in PBS without application of the sample solution (i.e., 0 ng/mL), indicated by a black solid line in Fig. 4b. Likewise, each measured point corresponding to one symbol is the average (i.e., Δλavg) of the five values of Δλ, obtained by five sequentially repeated spectral measurements, and the error bar of each point indicates the standard deviation of the five measured values. These measured points of Δλ were fitted using nonlinear regression based on Hill fitting with an adjusted R2 value of ~ 0.9995 in the case of the functionalized sensor head. The control experiment performed using the bare ST-NCF sensor head did not exhibit any remarkable changes for various sample concentrations, indicated by black triangles in Fig. 4c. Even at the highest sample concentration (i.e., 104 ng/mL), Δλavg was 0.005 nm, and the standard deviation of the measured Δλ values was ± 0.014 nm. This validates that SARS-CoV-2 spike protein does not specifically bind to the untreated ST-NCF sensor head. In contrast, when detecting the solution samples with concentrations of 1, 101, 102, 103, and 104 ng/mL using the functionalized ST-NCF sensor head, Δλavg values (including standard deviations) were evaluated as 0.072 ± 0.028, 0.176 ± 0.033, 1.66 ± 0.043, 2.23 ± 0.046, and 2.28 ± 0.039 nm, respectively. The LOD of the sensor head was estimated to be 8.49 ng/mL (blank + 3SD) for the detection of SARS-CoV-2 spike protein. It is noted that the wavelength shift of the indicator dip depends on the localized RI modulations of the surrounding medium, primarily caused by selective bioaffinity between ACE2 and SARS-CoV-2 spike protein. These results reveal that the bioconjugated ST-NCF sensor head has a sufficient capability to detect the selective binding of biological elements as an immunosensor. Table 1 presents the biosensing features of our platform and other previous works (i.e., different biosensors), including the bioreceptor, target, and LOD.
Detection of SARS-CoV-2 spike protein in saliva solution
After confirming the capability of the ACE2-immobilized ST-NCF sensor head to detect SARS-CoV-2 spike protein in PBS solution, we explored the effect of saliva existence in the course of the detection process addressed above. Saliva includes various organic components such as proteins, amino acids, or lipids, which can impair sensor performance including sensitivity deterioration. In this investigation, the discrimination capability of the sensor head for different concentrations of SARS-CoV-2 spike protein is assessed when the application of the saliva solution changes the measurement environment containing the target analytes. For this test, another biofunctionalized ST-NCF sensor head was fabricated under the same tapering condition as the previous sensor head used in the experiment of Fig. 4a was prepared. To begin with, before the sample (i.e., SARS-CoV-2 spike protein) injection into the container, we measured the transmission spectrum of the fabricated sensor head immersed in saliva solution. Next, the sensor head located in the container was exposed to a solution sample of 1 ng/mL in concentration for ~ 300 s and then washed with saliva. After the washing, the spectral measurement was carried out with the container filled with fresh saliva solution. This measurement sequence was repeated four times for the remaining sample concentrations (i.e., 101, 102, 103, and 104 ng/mL). In the mixed measurement environment, surplus unbound ACE2 receptors can be perturbed by numerous organic substances of saliva, affecting the surrounding medium RI of the ST-NCF sensor head. Figure 5a illustrates the bioaffinity binding-induced changes in the transmission spectrum of the sensor head, measured in saliva solution after the reaction with SARS-CoV-2 spike protein for every sample concentration (from 1 to 104 ng/mL). The inset of Fig. 5a shows the zoomed transmission spectrum observed over a wavelength range from 1510 to 1518 nm. Likewise, the transmission minimum was selected as the indicator dip. As in the case of the detection of SARS-CoV-2 spike protein in PBS solution, we could find that the indicator dip moved towards a longer wavelength region showing redshifts from 1513.96 to 1514.98 nm with increasing concentration of SARS-CoV-2 spike protein. Figure 5b shows the wavelength displacement Δλ of the indicator dip for each applied sample concentration, which was evaluated from the measured transmission spectra shown in Fig. 5a. The reference wavelength was also determined in the same way as used in the case of Fig. 4c. The measured points of Δλ were fitted by Hill-fitting-based regression again, and an adjusted R2 value of the fitted result was ~ 0.9882. In comparison with the Δλ data and the regression result shown in Fig. 4c, the fitted result of the measured data in Fig. 5b exhibited similar nonlinear behavior, although their signal contrast (i.e., the Δλ difference between concentrations) decreased. For the sample concentrations of 1, 101, 102, 103, and 104 ng/mL, Δλavg values (including standard deviations) were measured as 0.02 ± 0.017, 0.285 ± 0.032, 0.705 ± 0.037, 0.75 ± 0.033, and 0.84 ± 0.017 nm, respectively. The distinct wavelength shifts depending on the sample concentration corroborate that, even in the case of the existence of saliva, our ACE2-conjugated ST-NCF sensor head can still discern local RI changes caused by the bioaffinity binding between ACE2 and SARS-CoV-2 spike protein without severe corruption of the detection signal Δλ. In particular, the similarity between the regression results obtained in saliva and PBS solution also ensures the discrimination capability of our sensor head perturbed by saliva solution.
Selectivity and reproducibility
Owing to host specificity and viral infectivity, SARS-CoV-2 spike protein binds to ACE2 conjugated on the surface of the ST-NCF sensor head with bioaffinity. To investigate the cross-reactivity of our ACE2-conjugated ST-NCF sensor head, we compared its sensitivity to SARS-CoV-2 and MERS-CoV spike protein. For this comparison, we prepared the solution samples of MERS-CoV spike protein with concentrations of 1, 101, 102, 103, and 104 ng/mL and applied them to the sensor head. Before measurement of the transmission spectrum of the sensor head, the solution samples were injected into the container having the sensor head, and then the sensor head was washed with buffer solution (here, PBS or saliva solution) and was subsequently immersed in fresh buffer solution, as in the case of the detection process of SARS-CoV-2 spike protein. Blue and red squares in Fig. 5c show the indicator wavelength shifts (Δλ’s) induced by the reaction between two bioelements (i.e., MERS-CoV spike protein and ACE2), which were measured using PBS and saliva solution as measurement buffer, respectively. For a clear comparison, Δλ’s caused by the SARS-CoV-2 spike protein binding, measured using PBS (Fig. 4c) and saliva (Fig. 5b) solution, were also displayed with blue and red circles in Fig. 5c, respectively. All solid and dashed lines are the nonlinear regression results of measured Δλ data. For the sample concentrations of 1, 101, 102, 103, and 104 ng/mL, Δλavg values including standard deviations were evaluated as 0.005 ± 0.03, 0.1 ± 0.009, 0.12 ± 0.033, 0.18 ± 0.036, and 0.17 ± 0.078 nm with PBS solution, and 0.04 ± 0.01, 0.07 ± 0.024, 0.115 ± 0.030, 0.155 ± 0.017, and 0.2 ± 0.01 nm with saliva solution, respectively. In comparison with the detection result of SARS-CoV-2 spike protein, indicated by circular symbols, these measured Δλ values are not significant for sample concentrations higher than 10 ng/mL, which directly indicates the selectivity of our sensor head to SARS-CoV-2 spike protein. This can also be supported by the real-time transient monitoring of the indicator wavelength done before the post-bioreaction spectral measurement (performed after the binding reaction between ACE2 and MERS-CoV spike protein) like Fig. 5a. Fig. S2 represents the real-time wavelength variations of the indicator dip, measured when the sample concentration increases from 0 to 104 ng/mL. It can be observed from the figure that the indicator wavelength does not show a strong dependence on the sample concentration although it slightly increases with the sample concentration. The clear difference between Fig. S2 and Fig. 4a also suggests that our ACE2-conjugated ST-NCF sensor head has good specificity to distinguish the two biomolecules (i.e., SARS-CoV-2 and MERS-CoV spike protein).
Furthermore, to evaluate the reusability of the ST-NCF sensor head, which reflects its durability, and the reproducibility of the bioassay protocol, we repeated the experiment (specifically, the post-bioreaction spectral measurement) whose results were suggested in Fig. 5b employing the ST-NCF sensor head utilized in the previous experiment again. For this evaluation, the surface of the sensor head was functionalized with ACE2 again through the surface treatment process addressed in Sect. 2.3 starting from the piranha cleaning. Figure 5d shows the wavelength displacement Δλ of the indicator dip according to the sample concentration, which is obtained by combining three individual Δλ measurement results. These three experimental results presented in Fig. S3 and denoted by M-1, M-2, and M-3 were procured by using the same ST-NCF sensor head (i.e., one sensor head) and iterating the surface treatment process for every measurement, as shown in the inset of Fig. 5d. At a fixed sample concentration, each Δλ value in Fig. 5d was calculated by averaging three Δλ values at the corresponding sample concentration in the three plots of Fig. S3, and its error bar was determined by the standard deviation of these three Δλ values. At the highest sample concentration (i.e., 104 ng/mL), Δλ and its standard deviation were evaluated as about 0.97 and ± 0.18 nm, respectively. From the measurement results on Δλ, we can find that the Δλ behavior of Fig. 5d is very similar to that of Fig. 5b, which proves the reusability of our sensor head and the reproducibility of the bioassay protocol adopted here. In particular, this evaluation result of the reconstructed sensor head establishes its repeatability and reliability in immunosensing. Moreover, the reusability provides a preliminary basis for the development of a cost-effective biosensing platform.
Strengths and weaknesses
Optical fiber biosensors based on well-established fiber optic technology can provide an accurate biosensing solution that is tailorable to almost any clinical need and has relatively cheap and abundant material requirements [46]. Optica fiber as a component of the optical fiber sensors makes it especially effective in biological systems that may require ultra-sensitive detection of biological signals in hard-to-reach areas. These hard-to-reach areas include tissue or organs within the body where catheters or endoscopic tools can reach, places where there is a high risk of pathogenic or viral infection, and strong electromagnetic fields or high-energy radiation environments. Optical fiber biosensors are small enough to reach the hard-to-reach areas of the body and can also be adjusted from the general structure and adapted to more sensitive sensing in the body. In particular, they enable safe remote measurement thanks to electrical passivity, which does not require auxiliary power supplies for the sensor head, and low-loss/ultra-high-speed optical transmission characteristics, thereby fundamentally eliminating the risk of infection of measurers (e.g., medical or research personnel) in an environment susceptible to pathogenic or viral infection. On the other hand, compared to electronic biosensors, optical fiber biosensors have merits in electrical and fire safety owing to the use of glass waveguides and can operate without degradation of the sensor performance even in environments exposed to strong electromagnetic interference or noise. Moreover, they can guide light of different wavelengths simultaneously, giving them the ability to monitor and deliver information on more than one function at a time. For example, multiple detections of SARS-CoV-2 spike protein samples can be done through wavelength-division multiplexing using one optical fiber biosensor system. Furthermore, through some elaborate modifications of the sensor head structure and the sensor demodulation scheme, SARS-CoV-2 spike protein detection and ambient temperature monitoring can even be simultaneously carried out. In particular, our ST-NCF sensor head is not only relatively robust to lateral stress (compared with conventional TOF-based sensors) but also sensitive enough to target viral envelope proteins, let alone having all the advantages described above. In terms of the demerit of the proposed sensor, the optical spectrum analyzer used here for the spectral analysis is relatively costly in comparison with measurement instruments for electrical signals. However, this spectrum-analyzing instrument can be replaced with a much cheaper instrument like spectrometers or wavelength interrogators [47] and is expected to become cheaper continuously with the development of its constituent optical components. The other downside is that the sensitivity is not as high as that of an SPR optical fiber biosensor although it is comparable to that of conventional TOF-based sensors.
Conclusion
An ACE2-functionalized ST-NCF sensor head was employed for the label-free detection of SARS-CoV-2 spike protein for the first time. Slight tapering applied to NCF for sensitivity enhancement offers relative robustness to transverse stress, and the use of a standard fiber fusion splicer for the NCF tapering provides a simple and reproducible way of fabricating ST-NCF segments utilized as bare fiber sensor heads. Bare sensor heads were functionalized with human ACE2 bioreceptors through silanization and cross-linking processes. With the ACE2-conjugated ST-NCF sensor head, we investigated its capability of detecting SARS-CoV-2 spike protein samples in measurement buffers of PBS and saliva solutions by observing perturbation-induced wavelength shifts in the indicator dip. Through the real-time transient monitoring of the indicator dip, we could check the trend of the ongoing binding reaction between ACE2 and SARS-CoV-2 spike protein and confirmed that we could make rapid detection for all sample concentrations within 30 to 60 s. In particular, the cross-reactivity measurement results using MERS-CoV spike protein corroborate the selectivity of the sensor head to SARS-CoV-2 spike protein. Finally, we verified the reproducibility of the bioassay protocol adopted here by iterating the previous measurement processes done with saliva solution using the reconstructed ST-NCF sensor head. In addition, the cost of one sensor head unit (i.e., an ACE2-functionalized ST-NCF sensor head) was estimated as less than $20.9 by considering the costs of all materials utilized for its fabrication. The rapid detection capability, target specificity, reusability, and repeatability of our ST-NCF sensor head offer sufficient potential for a low-cost and portable optical fiber biosensor for early detection of SARS-CoV-2 spike protein. Furthermore, our fiber-optic diagnostic platform (i.e., ST-NCF transducer) based on biometric elements can expand its range of target analytes by employing appropriate receptors with target specificity.
References
Velavan TP, Meyer CG (2020) The COVID-19 epidemic. Trop Med Int Heal 25:278–280. https://doi.org/10.1111/tmi.13383
Yuki K, Fujiogi M, Koutsogiannaki S (2020) COVID-19 pathophysiology: a review. Clin Immunol 215:108427. https://doi.org/10.1016/j.clim.2020.108427
Conde Cardona G, Quintana Pájaro LD, Quintero Marzola ID, Ramos Villegas Y, Moscote Salazar LR (2020) Neurotropism of SARS-CoV 2: mechanisms and manifestations. J Neurol Ssci 412:116824. https://doi.org/10.1016/j.jns.2020.116824
Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46:586–590. https://doi.org/10.1007/s00134-020-05985-9
Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. https://doi.org/10.1038/s41586-020-2012-7
Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. https://doi.org/10.1038/s41564-020-0688-y
Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen Y-H, Zhang L, Wang X (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. https://doi.org/10.1038/cr.2013.92
Reguera J, Mudgal G, Santiago C, Casasnovas JM (2014) A structural view of coronavirus-receptor interactions. Virus Res 194:3–15. https://doi.org/10.1016/j.virusres.2014.10.005
Brielle ES, Schneidman-Duhovny D, Linial M (2020) The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses 12:497. https://doi.org/10.3390/v12050497
Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM (2020) SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol 10:1–17. https://doi.org/10.3389/fcimb.2020.587269
Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M (2020) Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol 27:1202–1208. https://doi.org/10.1038/s41594-020-00536-8
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. https://doi.org/10.1056/NEJMoa2001017
Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D (2020) Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 14:7617–7627. https://doi.org/10.1021/acsnano.0c03822
Zeng L, Li Y, Liu J, Guo L, Wang Z, Xu X, Song S, Hao C, Liu L, Xin M, Xu C (2020) Rapid, ultrasensitive and highly specific biosensor for the diagnosis of SARS-CoV-2 in clinical blood samples. Mater Chem Front 4:2000–2005. https://doi.org/10.1039/d0qm00294a
Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J (2020) Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 14:5268–5277. https://doi.org/10.1021/acsnano.0c02439
Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, Kim S-J, Lee JO, Kim BT, Park EC, Kim SI (2020) Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14:5135–5142. https://doi.org/10.1021/acsnano.0c02823
Xi H, Juhas M, Zhang Y (2020) G-quadruplex based biosensor: a potential tool for SARS-CoV-2 detection. Biosens Bioelectron 167:112494. https://doi.org/10.1016/j.bios.2020.112494
Zhang R, Wu J, Ao H, Fu J, Qiao B, Wu Q, Ju H (2021) A rolling circle-amplified G-quadruplex/hemin DNAzyme for chemiluminescence immunoassay of the SARS-CoV-2 protein. Anal Chem 93:9933–9938. https://doi.org/10.1021/acs.analchem.1c02229
Raziq A, Kidakova A, Boroznjak R, Reut J, Öpik A, Syritski V (2021) Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron 178:113029. https://doi.org/10.1016/j.bios.2021.113029
Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, Chaiyo S (2021) Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron 176:112912. https://doi.org/10.1016/j.bios.2020.112912
Esposito F, Srivastava A, Sansone L, Giordano M, Campopiano S, Iadicicco A (2021) Label-free biosensors based on long period fiber gratings: a review. IEEE Sens J 21:12692–12705. https://doi.org/10.1109/JSEN.2020.3025488
Zhao Y, Zhao J, Zhao Q (2020) Review of no-core optical fiber sensor and applications. Sens Actuators A Phys 313:112160. https://doi.org/10.1016/j.sna.2020.112160
Liu LL, Marques L, Correia R, Morgan SP, Lee SW, Tighe P, Fairclough L, Korposh S (2018) Highly sensitive label-free antibody detection using a long period fibre grating sensor Sens. Actuators B Chem 271:24–32. https://doi.org/10.1016/j.snb.2018.05.109
Janczuk-Richter M, Gromadzka B, Richter Ł, Panasiuk M, Zimmer K, Mikulic P, Bock WJ, Maćkowski S, Śmietana M, Jönsson JN (2020) Immunosensor based on long-period fiber gratings for detection of viruses causing gastroenteritis. Sensors 20:1–11. https://doi.org/10.3390/s20030813
Lee S-L, Kim J, Choi S, Han J, Seo G, Lee YW (2021) Fiber-optic label-free biosensor for SARS-CoV-2 spike protein detection using biofunctionalized long-period fiber grating. Talanta 235:122801. https://doi.org/10.1016/j.talanta.2021.122801
Hu W, Huang Y, Chen C, Liu Y, Guo T, Guan BO (2018) Highly sensitive detection of dopamine using a graphene functionalized plasmonic fiber-optic sensor with aptamer conformational amplification. Sens Actuators B Chem 264:440–447. https://doi.org/10.1016/j.snb.2018.03.005
Wang Q, Wang B-T (2018) Surface plasmon resonance biosensor based on graphene oxide/silver coated polymer cladding silica fiber. Sens Actuators B Chem 275:332–338. https://doi.org/10.1016/j.snb.2018.08.065
Wen H-Y, Huang C-W, Li Y-L, Chen J-L, Yeh Y-T, Chiang C-C (2020) A lamping U-shaped fiber biosensor detector for microRNA. Sensors 20:1509. https://doi.org/10.3390/s20051509
Tam TV, Hur SH, Chung JS, Choi WM (2021) Novel paper- and fiber optic-based fluorescent sensor for glucose detection using aniline-functionalized graphene quantum dots. Sens Actuators B Chem 329:129250. https://doi.org/10.1016/j.snb.2020.129250
Mustapha Kamil Y, Abu Bakar MH, Mustapa MA, Yaacob MH, Abidin NHZ, Syahir A, Lee HJ, Mahdi MA (2018) Label-free dengue E protein detection using a functionalized tapered optical fiber sensor. Sens Actuators B Chem 257:820–828. https://doi.org/10.1016/j.snb.2017.11.005
Chen L, Leng Y-K, Liu B, Liu J, Wan S-P, Wu T, Yuan J, Shao L, Gu G, Fu YQ, Xu H, Xiong Y, He X-D, Wu Q (2020) Ultrahigh-sensitivity label-free optical fiber biosensor based on a tapered singlemode-no core-singlemode coupler for Staphylococcus aureus detection. Sens Actuators B Chem 320:128283. https://doi.org/10.1016/j.snb.2020.128283
Ravikumar R, Chen LH, Jayaraman P, Poh CL, Chan CC (2018) Chitosan-nickel film based interferometric optical fiber sensor for label-free detection of histidine tagged proteins. Biosens Bioelectron 99:578–585. https://doi.org/10.1016/j.bios.2017.08.012
Yang W, Zhang S, Geng T, Li L, Li G, Gong Y, Zhang K, Tong C, Lu C, Sun W, Yuan L (2019) High sensitivity refractometer based on a tapered-single mode-no core-single mode fiber structure. Sensors 19:1722. https://doi.org/10.3390/s19071722
Xu W, Shi J, Yang X, Xu D, Rong F, Zhao J, Yao J (2017) Improved numerical calculation of the single-mode-no-core-single-mode fiber structure using the fields far from cutoff approximation. Sensors 17:2240. https://doi.org/10.3390/s17102240
Sheu F-W, Huang Y-S (2013) Trapping and propelling microparticles at long range by using an entirely stripped and slightly tapered no-core optical fiber. Sensors 13:2884–2894. https://doi.org/10.3390/s130302884
Xiong W, Ambichl P, Bromberg Y, Redding B, Rotter S, Cao H (2016) Spatiotemporal of light transmission through a multimode fiber with strong mode coupling Phys. Rev Lett 117:053901. https://doi.org/10.1103/PhysRevLett.117.053901
Zhang M, Zhu G, Lu L, Lou X, Zhu L (2019) Refractive index sensor based on ultrafine tapered single-mode nocladding single-mode fiber structure. Opt Fiber Technol 48:297–302. https://doi.org/10.1016/j.yofte.2019.01.008
Wang Q, Farrell G, Yan W (2016) Investigation on single-mode-multimode-single-mode fiber structure. J Lightwave Technol 26:12–519. https://doi.org/10.1109/JLT.2007.915205
Taha BA, Ali N, Sapiee NM, Fadhel MM, Yeh RMM, Bachok NN, Mashhadany YA, Arsad N (2021) Comprehensive review tapered optical fiber configurations for sensing application: trend and challenges. Biosensors 11:253. https://doi.org/10.3390/bios11080253
Zainuddin NH, Chee HY, Ahmad MZ, Mahdi MA, Bakar MHA, Yaacob MH (2018) Sensitive Leptospira DNA detection using tapered optical fiber sensor. J Biophotonics 11:e201700363. https://doi.org/10.1002/jbio.201700363
Lin H-Y, Huang C-H, Cheng G-L, Chen N-K, Chui H-C (2012) Tapered optical fiber sensor based on localized surface plasmon resonance. Opt Express 20:21693–21701. https://doi.org/10.1364/oe.20.021693
Kaushik S, Pandey A, Tiwari UK, Sinha RK (2018) A label-free fiber optic biosensor for Salmonella Typhimurium detection. Opt Fiber Technol 46:95–103. https://doi.org/10.1016/j.yofte.2018.09.012
Herrera ND, Esteban Ó, Navarrete M-C, González-Cano A, Benito-Peña E, Orellana G (2011) Improved performance of SPR sensors by a chemical etching of tapered optical fibers. Opt Lasers Eng 49:1065–1068. https://doi.org/10.1016/j.optlaseng.2011.03.012
Zibaii MI, Latifi H, Asadollahi A, Bayat AH, Dargahi L, Haghparast A (2016) Label free fiber optic apta-biosensor for in-vitro detection of dopamine. J Lightwave Technol 34:4516–4524. https://doi.org/10.1109/JLT.2016.2550533
Tian Y, Wang W, Wu N, Zou X, Wang X (2011) Tapered optical fiber sensor for label-free detection of biomolecules. Sensors 11:3780–3790. https://doi.org/10.3390/s110403780
Wang X-D, Wolfbeis OS (2020) Fiber-optic chemical sensors and biosensors (2015–2019). Anal Chem 92:397–430. https://doi.org/10.1021/acs.analchem.9b04708
Waltermann C, Guehlke P, Koch J, Schippers W (2019) Shaping spectra within optical fibers: embedding high-resolution spectrometers into the cores of optical fibers. Opt Meas Technol 2:42–45. https://doi.org/10.1002/phvs.201900013
Acknowledgements
This research was supported by the Korea Institute for Advancement of Technology (KIAT) grant funded by the Korean Government (MOTIE) (P0012451, The Competency Development Program for Industry Specialist), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1I1A3A01046232), and National Research Council of Science and Technology funded by the Ministry of Science and ICT, Republic of Korea (No. CRC‐16‐01‐KRICT).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, J., Lee, SL., Kim, J. et al. SARS-CoV-2 spike protein detection using slightly tapered no-core fiber-based optical transducer. Microchim Acta 189, 321 (2022). https://doi.org/10.1007/s00604-022-05413-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05413-3