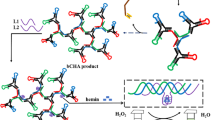
Abstract
The spread of African swine fever virus (ASFV) caused huge economic costs, so early detection is particularly important. Here, we established a fluorescence biosensor based on carbon nanodots (CNDs) and loop-mediated isothermal amplification (LAMP) to ultra-sensitively detect ASFV. LAMP with high efficiency produced a large amount of pyro phosphoric acid and caused pH change in a short time. CNDs with strong light stability had a large fluorescence response at the emission wavelength of 585.5 nm to small pH change by the excitation wavelength of 550 nm. The biosensor realized “turn-off–on” mode for ASFV detection with the detection limit as low as 15.21 copies μL−1. In addition, the biosensor had high accuracy in the actual sample assay. Therefore, the biosensor achieved rapid, sensitive, low-cost, and simple detection for ASFV. Moreover, the biosensor broadened the detection pathway of LAMP as a tool with great development prospect.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
African swine fever (ASF) has been listed as a notifiable disease by the World Organization for Animal Health [1]. ASF is caused by the African swine fever virus (ASFV), which is a highly stable, lethal, and contagious double-stranded DNA virus [2]. According to the different susceptibility of virus strains and hosts, clinical symptoms of ASFV can be a chronic disease, hemorrhagic fever, and hyperacute death [3, 4]. However, there is currently no commercially available vaccine for ASFV, leading to affecting domestic and wild pigs of all breeds and ages, causing mortality rates approaching 100% [5, 6]. It causes great harm to the pig industry with no sign of slowing down and destroys the stability of the global economic market [7]. Therefore, early detections and rapid diagnosis are essential for the prevention and monitoring of animal infection with ASFV.
The detection methods of ASFV can be divided into immunological detection methods and molecular biological detection methods. Immunological detection methods include blood adsorption test (HAD) [8], enzyme-linked immunosorbent assay (ELISA) [9], fluorescent antibody test (FAT) [10], and lateral flow assay (LFA) [11]. However, these methods are not suitable for early diagnosis because they are time-consuming and have low sensitivity and hysteresis. Therefore, nucleic acid detection plays an important role in the early diagnosis of ASFV. As the common nucleic acid detection methods, conventional polymerase chain reaction (PCR) and quantitative PCR (qPCR) [12] require proficiency in personnel operation and expensive instruments. In contrast, isothermal amplification is easier to operate and costs less time, such as recombinant polymerase amplification (RPA) [13], loop-mediated isothermal amplification (LAMP) [14], and hybrid chain reaction (HCR) [15], rolling circle amplification (RCA) [16], and strand displacement amplification (SDA) [17]. Among them, LAMP relies on Bst DNA polymerase and four primers to detect nucleic acid at a constant temperature of 60–65 ℃ with high specificity and sensitivity. It is more suitable for non-professional or mobile laboratories. Moreover, LAMP has more terminal detection methods, such as turbidimetry and colorimetry [18]. During DNA synthesis, a large amount of pyrophosphoric acid (PPi) is produced, forming white precipitates with Mg2+ and being detected by turbidimetry [19]. However, the turbidity caused by low concentration targets is difficult to be detected. In addition, PPi produced by LAMP can cause pH changes in the solution. Some acid–base indicators, such as neutral red [1], hydroxy naphthol blue (HNB) [20], calcein [21], and SYBR green [22], can be used to distinguish positive about the negative, but these indicators require a wide range of pH. However, the reaction system of LAMP is only 25 µl. Especially for the target with a small concentration, the pH change generated by LAMP is small, so a more sensitive indicator for pH is urgently needed.
Carbon nanodots (CNDs) are spherical fluorescent material less than 10 nm in diameters, which has a good solubility in water, adjustable photoluminescence (fluorescent), low toxicity, good biocompatibility, and stable chemical properties [23]. Due to their unique photoelectric properties, CNDs have shown great application prospects in photocatalysis, detection of heavy metal ions and organic molecules, biological imaging, and other fields [23,24,25]. More importantly, some CNDs respond significantly to pH changes, and CNDs have great potential as pH indicators [26].
To solve the current methods with low sensitivity by pH changes of LAMP reaction, here, we constructed a biosensor based on the CNDs and LAMP to detect the small concentration of ASFV. Since the synthesized CNDs had a large response to small pH changes from 8.3 to 8.9, the biosensor constructed a turn-off–on mode to amplify signals through the fluorescence response of CNDs before and after LAMP reaction. And the biosensor realized the ultra-sensitive detection of ASFV with a detection limit as low as 15.21 copies μL−1. In addition, CND synthesis was low-cost and stable, and LAMP reaction can be completed in a short time under constant temperature, which realized simple, rapid, and low-cost nucleic acid detection and was beneficial to point-of-care test (POCT). Furthermore, this method expanded the detection pathway of LAMP and also improved the sensitivity of LAMP by the signal amplification of CNDs.
Material and methods
Reagents and instruments
All nucleic acids used in this study were offered by Sangon Biotech (Shanghai, China) and the sequences are showed in Table S1. Classical swine fever virus (CSFV) and porcine circovirus (PCV) were offered by Chongqing customs technology center. The nucleic acids of pseudorabies virus (PRV) and Peste des petits ruminant virus (PPRV) were gifted by Dr. Ai from Kunming Customs Technology Center. Citric acid and 5-aminosalicylic acid were purchased from Macklin (Shanghai, China). QIAamp DNA Mini Kit was purchased from QIAGEN, Germany. Bst DNA polymerase, 10 × isothermal amplification buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH = 8.8), and 100 mM MgSO4 were purchased from Eiken Chemical, Japan. DL500 and DL2000 DNA Marker and 2 × Premix Ex Taq (Probe qPCR) were purchased from TaKaRa. ABI 7500 Fast fluorescence quantitative PCR instrument was purchased from life technology TM, USA. Gel imager bought from Bio Rad Co., Ltd. X-ray photoelectron spectroscopy (XPS, XRD-7000) was purchased from JEOL (Japan) for characterizing the elements and their chemical states of CNDs. Transmission electron microscopy (TEM, JEOL Ltd., Japan) was used in morphology characterization. The zeta potential data was obtained by using a Zetasizer nano (Malvern, England).
Synthesis of CNDs
The hydrothermal method is the preferred method for the green synthesis of CNDs according to the previous studies [27]. And the detailed steps of CNDs synthesis are described in Section S1.
Characterization performance of CNDs
The micromorphology of the CND was characterized by TEM. Its chemical composition and crystal structure were characterized by X-ray photoelectron spectroscopy (XPS, Al Kα excitation light source). The zeta potential was used to detect the potential of CNDs in the buffer with different pH.
ASFV assay by the biosensor
The P72 gene of ASFV was used as the target. The 25-µL LAMP reaction system consisted of 1 µL of 5 µM F3 and B3 primers, 1 µL of 20 µM FIP and BIP primers, 1.5 µL of 10 mM MgSO4, 8 U Bst DNA polymerase, 2 µL sample, 3.5 µL 10 mM dNTP and 13 µL buffer (10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4). It was incubated at 62 ℃ for 30 min. After then, 75 µL CNDs diluted 30 times was added into the LAMP system at room temperature for 2 min to measure fluorescence by LS-55 spectrophotofluorometer (P. E. USA) with the exciting spectrum of 550 nm.
qPCR for detecting ASFV
As a standard method, qPCR verified the accuracy of the method-based CNDs and LAMP by testing actual samples. Sequences of primers and probes are presented in Table S1 (Supporting information). The nucleic acids of samples were extracted according to the instruction of QIAamp DNA Mini Kit. The qPCR system contained 1 µL of 10 µM SN/T-F and SN/T-R primers, 1 µL of 10 µM SN/T-probe, 12.5 µL of 2 × Premix Ex Taq (Probe qPCR), 4.5 µL ddH2O, and 5 µL of sample nucleic acid. The system was programmed: 95 ℃ for 5 min, followed by 40 cycles, including 95 ℃ for 10 s and 58 ℃ for 34 s.
Agarose gel electrophoresis
Gel electrophoresis was used to verify the feasibility of the method. The amplified product was mixed with 6 × loading buffer for 2% agarose gel at 130 V for 20 min.
Real samples analysis by the biosensor
The nucleic acid was extracted and purified from blood or tissue according to the instruction of the QIAamp DNA Mini Kit. And 2 µL of nucleic acid was added into the LAMP system at 62 ℃ for 30 min. Next, 75 µL of diluted 30 times CNDs was added and placed at room temperature for 2 min for measuring fluorescence intensity by LS-55 spectrophotofluorometer with the exciting spectrum of 550 nm.
Results and discussion
Characterization of the CNDs
TEM was used to characterize the morphology of CNDs. Figure 1a showed that the CNDs were spherical and evenly distributed in the aqueous solution without obvious aggregation. Their sizes ranged from 2.01 to 4.40 nm, with an average diameter of 3.41 ± 0.5 nm. In addition, the high-resolution TEM image revealed that the CNDs had obvious lattice fringes of 0.204 nm. This indicated that the synthesized CNDs had uniform size and good dispersion. However, when the CNDs were added into the buffer with a pH of 8.50, an obvious aggregation phenomenon was shown in Fig. 1b. Their diameters were distributed from 21.37 to 23.45 nm, and the mean diameter was 23.286 nm. It illustrated that the CNDs had an obvious response to the change of pH. Moreover, when CNDs with the negative electricity were added to buffers with different pH, their potential was different in Fig. 1c. With the increase of pH, the potential decreased continuously, indicating that the CNDs had a significant response to pH. Furthermore, different excitation wavelengths were used to verify the best excitation wavelength of the CNDs. As shown in Fig. 1d, the fluorescence intensity increased with the increase of excitation wavelength from 500 to 560 nm. And the emission wavelengths of the CNDs were all at 585.5 nm without deviation. However, the emission wavelength was too close to the excitation wavelength at 560 nm, which could cause interference. Therefore, 550 nm was selected as the best excitation wavelength. Furthermore, XPS was used to verify the composition of the elements of the CNDs. In Fig. 2a, there were three distinct peaks 528.51, 396.82, and 282.16, corresponding to O1s, N1s, and C1s. This indicated that the main components of CNDs were C, N, and O. As shown in Fig. 2b, C1s XPS spectrogram exhibited the obvious peaks at 284.70, 285.98, and 288.34, representing C–C/C = C, C–N, and C = O respectively. The N1s XPS spectrogram (Fig. 2c) was fitted with three functional groups including C = N–C, C–N, N–H, which correspond respectively with 398.90, 399.70, and 400.70 eV. Figure 2d displayed that O1s XPS spectrogram can be divided into six peaks at 531.22, 531.50, 531.68, 532.10, 532.60, and 533.40, representing C = O, COOH, C–N–O, C–OH, O–C–O, and C–O. The above results proved that the CNDs contained abundant hydroxyl and carboxyl active groups on the surfaces, which are sensitive to pH.
a represents TEM image of CNDs, the enlarged TEM image, and the size distribution diagram of CNDs. b shows the TEM image of the distribution of CNDs in the buffer (pH = 8.50). c manifested the zeta potential CNDs in the buffer with different pH. d indicates the fluorescence spectrum of CNDs at different excitation wavelengths from 500 to 560 nm
Principle of ASFV detection by CNDs combined with LAMP
Scheme 1 showed the principle of CNDs combined with LAMP for ASFV detection. LAMP is a sensitive and specific amplification method that rapidly generates a large amount of DNA under isothermal conditions. Under the action of four primers for calibrating the target with high specificity, ASFV is amplified by Bst DNA polymerase to produce DNA. In this process, dNTP is broken down into deoxynucleotide (dNMP) and pyrophosphoric acid (PPi), where dNMP is involved in DNA synthesis, and PPi is released into the buffer. The PPi could change the pH of the buffer.
On the other hand, the CNDs synthesized in this paper will aggregate with the increase of pH. That is, when excitation wavelength hits the CNDs, they absorb photons, making an electronic transition and fluorescing as it travels back to the valence electrons. When pH increases, the hydroxyl and carboxyl groups on the surface of the CNDs are deprotonated and the electron supply capacity of oxygen is improved. This enhances the hydrogen bond interaction between CNDs to aggregate and thickens the electron trap, which enables photons to be emitted in a non-radiative form, thereby quenching fluorescence. Therefore, the CND realizes the “turn-off” mode as the pH increases. When the CNDs are added to the alkaline buffer, the fluorescence will be quenched.
LAMP can reduce pH due to generating a large amount of PPi, causing CNDs in the buffer to disperse. Therefore, the fluorescence is enhanced, and it realizes the “turn-on” mode as the target increases. In other words, the biosensor constructed by LAMP and CNDs achieves highly sensitive detection of ASFV through the turn-off–on mode.
The turn-off mode of response of CNDs to pH
In theory, with the increase of pH, active groups on the surface of CNDs were deprotonated, resulting in aggregation and decreasing fluorescence intensity. In fact, Tris–HCl at different pH was employed to verify the performance of CNDs. The fluorescence intensity decreased with pH increasing from 8.32 to 9.01 in Fig. 3a. Moreover, the linear fitting of the fluorescence intensity and pH value ranged from 8.32 to 9.01 showed a good linear relationship in Fig. 3b, F= − 813.76 pH + 7725.41, R2 = 0.9725. Since Bst DNA polymerase of LAMP reaction has the high activity in a weak alkaline environment, the buffers (10 mM (NH4)2SO4, 50 mM KCl, and 2 mM MgSO4) of different pH values were prepared. As shown in Fig. 3c, as the pH value of the buffer increased, the fluorescence intensity decreased, which had the same trend as that of Tris–HCl. The pH value from 8.3 to 8.9 and fluorescence intensity was linearly fitted in Fig. 3d, and a good linear relationship was obtained, F = − 1401.23 pH + 12,620.04, R2 = 0.9756. Although the response range of CNDs to pH was small, a small pH change in an alkaline environment can cause a large change in fluorescence intensity. This was an important feature for the LAMP reaction. In other words, the volume of the LAMP reaction system was small, and the pH change caused by the reaction was small. More importantly, the fluorescence intensity of CNDs was not affected by temperature (in Section S2). So the biosensor can ultra-sensitively detect ASFV in small concentrations by LAMP and the CNDs.
Characterization of the biosensor for ASFV detection
The biosensor was used to detect small concentrations of ASFV under optimal conditions (in Section S3). In Fig. 4a, as the concentrations of ASFV increased from 50 to 250 copies μL−1, the fluorescence intensity increased. This manifested more targets caused LAMP reactions, resulting in large pH changes. The CNDs were more dispersed and produced more fluorescence. There was a good linear relationship between 50 and 250 copies μL−1, ΔF = 0.669·C + 51.612, R2 = 0.9725 in Fig. 4b. The detection limit was as low as 15.21 copies μL−1 according to the 3σ rule. In addition, agarose gel electrophoresis was used to verify LAMP amplification products of targets with different concentrations. It was found that the brightness of the bands decreased significantly with the decrease of concentration in Fig. 4c. At concentration as low as 50 copies μL−1, there were no visible bands. By contrast, the biosensor based on CNDs had higher sensitivity to detect ASFV. Compared with other methods, the biosensor had higher sensitivity to detect ASFV (Table S2).
a showed the fluorescence intensity of different concentrations of ASFV. b exhibited the linear relationship between the ΔF at the emission wavelength of 585.5 nm and concentration from 50 to 250 copies μL−1. c demonstrated the agarose gel results of products with different concentrations of ASFV. d indicated the specificity of the biosensor with the ASFV of 250 copies μL−1 at the emission wavelength of 585.5 nm
Specificity of biosensor for ASFV detection
To verify the specificity of the biosensor, viruses commonly occurring in pigs were selected as templates with the same concentration of 250 copies μL−1, for example, CSFV, PRV, PCV, and PRRSV. As shown in Fig. 4d, the fluorescence intensity at the emission wavelength of 585.5 nm of other viruses was the same as that of the control group, while ASFV displayed obvious strong fluorescence, indicating that the LAMP reaction had strong specificity. In addition, agarose gel electrophoresis was used to verify the specificity of LAMP in Fig. S1. It was found that only ASFV had obvious gradient bands, while others had no band. The results were consistent with those of detection by the biosensor, manifesting that CNDs combined with LAMP was specific for ASFV assay.
Reproducibility and repeatability of the biosensor
Reproducibility and repeatability were important parameters of the biosensor. Repeated experiments of the biosensor for detecting ASFV of 200 copies μL−1 by different researchers showed that RSD was 3.64% in Fig. 5a, demonstrating that CNDs combined with LAMP had good reproducibility for ASFV detection. In addition, the experiment for ASFV of 200 copies μL−1 was repeated six times in Fig. 5b, and the RSD was 2.11%, indicating that the biosensor had good repeatability.
Detection of ASFV in the real samples
Seventeen samples from tissue, blood of pigs, and meat products were used to verify the performance of the biosensor. It was found that the fluorescence intensity of 3 samples was similar to that of the negative control (NC), while the remaining 14 samples had strong fluorescence in Fig. 6a. That manifested there were 3 negative samples and 14 positive samples. Moreover, qPCR as the standard method was used to verify accuracy. As shown in Fig. 6b, Ct values of 14 positive samples were significantly less than 35 cycles, while 3 negative samples had no fluorescence. These results were consistent with the biosensor, indicating that the biosensor showed high accuracy in actual samples. Compared with qPCR, CNDs binding LAMP have better advantages, such as no dependence on expensive large instruments to accurately control the temperature, easy synthesis, and cheap. In addition, agarose gel electrophoresis was used as an auxiliary tool for verification. The results showed that 14 samples had obvious bands, and 3 samples had no bands in Fig. S2. These results were consistent with those of the biosensor. In addition, targets at different concentrations were added to the serum of pigs diluted 20 times to be detected by the biosensor. Section S4 showed that the recovery rate was between 97.57% and 105.42% in Table S3. Those proved that the CND combined LAMP has a good ability to detect real samples.
Conclusions
To ultra-sensitively detect ASFV, a fluorescence biosensor was built based on the LAMP and CNDs. Through the large response of CNDs to pH before and after LAMP reaction, the biosensor constructed the “turn-off–on” mode. The biosensor had more advantages in practical applications. First of all, LAMP had high amplification efficiency under isothermal conditions in a short time, which got rid of the dependence of large instruments. In addition, the CNDs were easy to synthesize, green, and had a large response to pH changes, which was conducive to the promotion of the biosensor. At the same time, the biosensor based on pH assay solved the problem of aerosol pollution compared with method-based LAMP combined with fluorescent dyes or agarose gel electrophoresis. Taking full advantage of LAMP and CNDs, the biosensor achieved fast and sensitive detection in 32 min. More importantly, the biosensor was easier to operate and cheaper, which was more suitable for POCT. Above all, the biosensor was a potential tool for ASFV detection. In the future, more sensitive to pH and more stable fluorescence materials are expected to apply for the biosensor and lateral flow dipstick. In addition, we are looking forward to LAMP reaction system that does not require nucleic acid extraction and has higher efficiency, which can save a lot of labor and time.
References
Wang Y, Dai JF, Liu YS, Yang JF, Hou Q, Ou YW, Ding YZ, Ma B, Chen HT, Li MM, Sun YF, Zheng HX, Zhang K, Wubshet AK, Zaberezhny AD, Aliper TI, Tarasiuk K, Pejsak Z, Liu ZJ, Zhang YG, Zhang J (2021) Development of a potential penside colorimetric LAMP assay using neutral red for detection of African swine fever virus. Front Microbiol 12:609821
Zhu YS, Shao N, Chen JW, Qi WB, Li Y, Liu P, Chen YJ, Bian SY, Zhang Y, Tao SC (2020) Multiplex and visual detection of African swine fever virus (ASFV) based on Hive-Chip and direct loop-mediated isothermal amplification. Anal Chim Acta 1140:30–40
Blome S, Gabriel C, Beer M (2013) Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res 173:122–130
Costard S, Mur L, Lubroth J, Sanchez-Vizcaino JM, Pfeiffer DU (2013) Epidemiology of African swine fever virus. Virus Res 173:191–197
Borca MV, Ramirez-Medina E, Silva E, Vuono E, Rai A, Pruitt S, Espinoza N, Velazquez-Salinas L, Gay CG, Gladue DP (2021) ASFV-G- increment I177L as an effective oral nasal vaccine against the Eurasia strain of Africa swine fever. Viruses-Basel 13(5)
Lopera-Madrid J, Medina-Magues LG, Gladue DP, Borca MV, Osorio JE (2021) Optimization in the expression of ASFV proteins for the development of subunit vaccines using poxviruses as delivery vectors. Sci Rep-Uk 11(1)
Gaudreault NN, Madden DW, Wilson WC, Trujillo JD, Richt JA (2020) African Swine fever virus: an emerging DNA arbovirus. Front Vet Sci 7
Rodriguez JM, Yanez RJ, Almazan F, Vinuela E, Rodriguez JF (1993) African swine fever virus encodes a Cd2 homolog responsible for the adhesion of erythrocytes to infected-cells. J Virol 67:5312–5320
Cubillos C, Gomez-Sebastian S, Moreno N, Nunez MC, Mulumba-Mfumu LK, Quembo CJ, Heath L, Etter EMC, Jori F, Escribano JM, Blanco E (2013) African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res 173:159–167
Gallardo C, Nieto R, Soler A, Pelayo V, Fernandez-Pinero J, Markowska-Daniel I, Pridotkas G, Nurmoja I, Granta R, Simon A, Perez C, Martin E, Fernandez-Pacheco P, Arias M (2015) Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in Eastern European Union countries: how to improve surveillance and control programs. J Clin Microbiol 53:2555–2565
Sastre P, Gallardo C, Monedero A, Ruiz T, Arias M, Sanz A, Rueda P (2016) Development of a novel lateral flow assay for detection of African swine fever in blood. Bmc Vet Res 12
Zsak L, Borca MV, Risatti GR, Zsak A, French RA, Lu Z, Kutish GF, Neilan JG, Callahan JD, Nelson WM, Rock DL (2005) Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J Clin Microbiol 43:112–119
Miao FM, Zhang JY, Li N, Chen T, Wang LD, Zhang F, Mi LJ, Zhang JX, Wang SC, Wang Y, Zhou XT, Zhang YY, Li M, Zhang SF, Hu RL (2019) Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African swine fever virus. Front Microbiol 10:1004
Wang DG, Yu JH, Wang YZ, Zhang M, Li P, Liu M, Liu YH (2020) Development of a real-time loop-mediated isothermal amplification (LAMP) assay and visual LAMP assay for detection of African swine fever virus (ASFV). J Virol Methods 276:113775
Li L, Jiang HS, Meng XX, Wen XH, Guo QP, Li ZH, Wang J, Ren YZ, Wang KM (2021) Highly sensitive detection of cancer cells via split aptamer mediated proximity-induced hybridization chain reaction. Talanta 223:121724
Meng YR, Zhang D, Zou X, Ma F, Kang Q, Zhang CY (2021) A trifunctional split dumbbell probe coupled with ligation-triggered isothermal rolling circle amplification for label-free and sensitive detection of nicotinamide adenine dinucleotide. Talanta 224:121962
Zhang HG, Li FY, Wang LL, Shao S, Chen HL, Chen XG (2020) Sensitive homogeneous fluorescent detection of DNA glycosylase by target-triggering ligation-dependent tricyclic cascade amplification. Talanta 220:121422
Shi YQ, Xu M, Duan XQ, Li SL, Ding JW, Chen LM (2021) WarmStart colorimetric loop-mediated isothermal amplification for the one-tube, contamination-free and visualization detection of Shigella flexneri. Int J Infect Dis 112:55–62
Arunrut N, Suebsing R, Withyachumnarnkul B, Kiatpathomchai W (2014) Demonstration of a very inexpensive, turbidimetric, real-time, RT-LAMP detection platform using shrimp Laem-Singh virus (LSNV) as a Model. Plos One 9(9):e108047
Goto M, Honda E, Ogura A, Nomoto A, Hanaki KI (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167-+
Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K (2005) Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol 43:2895–2903
Hill J, Beriwal S, Chandra I, Paul VK, Kapil A, Singh T, Wadowsky RM, Singh V, Goyal A, Jahnukainen T, Johnson JR, Tarr PI, Vats A (2008) Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J Clin Microbiol 46:2800–2804
Kong DP, Yan FY, Shi DC, Ye QH, Han ZY, Chen L, Wang L (2015) Carbon dots: synthetic methods and applications as fluorescent probes for the detection of metal ions, inorganic anions and organic molecules. J Iran Chem Soc 12:1841–1857
Tan L, He R, Chen KC, Peng RF, Huang C, Yang R, Tang YW (2016) Ultra-high performance liquid chromatography combined with mass spectrometry for determination of aflatoxins using dummy molecularly imprinted polymers deposited on silica-coated magnetic nanoparticles. Microchim Acta 183:1469–1477
Yang XM, Zhuo Y, Zhu SS, Luo YW, Feng YJ, Dou Y (2014) Novel and green synthesis of high-fluorescent carbon dots originated from honey for sensing and imaging. Biosens Bioelectron 60:292–298
Susumu K, Mei BC, Mattoussi H (2009) Multifunctional ligands based on dihydrolipoic acid and polyethylene glycol to promote biocompatibility of quantum dots. Nat Protoc 4:424–436
Shi LH, Bao YJ, Zhang Y, Zhang CH, Zhang GM, Dong C, Shuang SM (2021) Orange emissive carbon nanodots for fluorescent and colorimetric bimodal discrimination of Cu2+ and pH. Analyst 146:1907–1914
Funding
This work was supported by the National Natural Science Foundation of China (NO.81772290), Chongqing science and technology commission (cstc2021jcyj-msxmX0608), Graduate Scientific Research and Innovation Foundation of Chongqing, China (Grant CYB21070), Chongqing Technology Innovation and Application Development Special Project (CSTC2019jscx-gksbX0132), Research Project of General Administration of Customs (2019HK029), Research Project of Chongqing Customs(2020CQKY10), Chongqing Graduate Tutor Team Construction Project, Analytical and Testing Center of Chongqing University for TEM and the sharing fund of Chongqing University’s large equipment.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, G., Qiu, Y., Long, K. et al. Carbon nanodots combined with loop-mediated isothermal amplification (LAMP) for detection of African swine fever virus (ASFV). Microchim Acta 189, 342 (2022). https://doi.org/10.1007/s00604-022-05390-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05390-7