Abstract
N6-Methyladenosine (m6A) is the most abundant RNA modification in eukaryotic messenger RNA (mRNA). A highly sensitive electrochemical immunosensor is described for the determination of m6A-RNA. The method is based on the use of antibody (anti-m6A) and PtCo mesoporous nanospheres (MPNs). The analogously modified probe of type m6A-DNA-PtCo competes with m6A-RNA for antibodies on the gold electrode as an electrical signal probe. The electrical signal, best acquired at a working potential of −0.37 V (vs. Ag/AgCl) reflects the concentration of m6A. The PtCo MPNs catalyze the reduction of H2O2, and this amplifies the current and enhances sensitivity. The detection time of the assay is <1.5 h. Under optimal conditions, response is linear in the 0.005 to 100 nM m6A RNA concentration range, and the detection limit is 2.1 pM. The results obtained by this immunoassay with human cell lines are comparable to those obtained with a commercial kit.

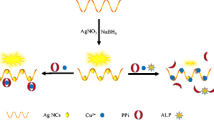
Schematic representation of a method for electrochemical determination of m6A-modified mRNA. Anti-m6A Ab: antibody against m6A; BSA: bovine serum albumin; PtCo: PtCo mesoporous nanospheres; SH-m6A-DNA: DNA modified with both m6A and thiol groups; DPV: differential pulse voltammetry.





Similar content being viewed by others
References
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM (2018) RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67(6):2254–2270. https://doi.org/10.1002/hep.29683
Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, Jia G (2018) An elongation and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-methyladenosine modifications. Angew Chem Int Ed 57(49):15995–16000. https://doi.org/10.1002/anie.201807942
Louloupi A, Ntini E, Conrad T, Ørom UAV (2018) Transient N-6-Methyladenosine Transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep 23(12):3429–3437. https://doi.org/10.1016/j.celrep.2018.05.077
Zhao BS, Roundtree IA, He C (2016) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18(1):31–42. https://doi.org/10.1038/nrm.2016.132
Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR (2016) Reversible methylation of m6Am in the 5’cap controls mRNA stability. Nature 541(7637):371–375. https://doi.org/10.1038/nature21022
Frye M, Harada BT, Behm M, He C (2018) RNA modifications modulate gene expression during development. Science 361:1346–1349. https://doi.org/10.1126/science.aau1646
Hsu PJ, Shi H, He C (2017) Epitranscriptomic influences on development and disease. Genome Biol 18(1):197. https://doi.org/10.1186/s13059-017-1336-6
Vu LP, Cheng Y, Kharas MG (2019) The biology of m6A RNA methylation in Normal and malignant hematopoiesis. Cancer Discov 9(1):25–33. https://doi.org/10.1158/2159-8290.CD-18-0959
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C (2011) N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7(12):885–887. https://doi.org/10.1038/nchembio.687
Yan M, Wang Y, Hu Y, Feng Y, Dai C, Wu J, Wu D, Zhang F, Zhai Q (2013) A high-throughput quantitative approach reveals more small RNA modifications in mouse liver and their correlation with diabetes. Anal Chem 85(24):12173–12181. https://doi.org/10.1021/ac4036026
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR (2015) Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12(8):767–772. https://doi.org/10.1038/nmeth.3453
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’UTRs and near stop codons. Cell 149(7):1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Yin D, Tao Y, Tang L, Li W, Zhang Z, Li J, Xie G (2017) Cascade toehold-mediated strand displacement along with non-enzymatic target recycling amplification for the electrochemical determination of the HIV-1 related gene. Microchim Acta 184(10):3721–3728. https://doi.org/10.1007/s00604-017-2368-z
Wang F, Chu Y, Ai Y, Chen L, Gao F (2019) Graphene oxide with in-situ grown Prussian blue as an electrochemical probe for microRNA-122. Microchim Acta 186(2):116–119. https://doi.org/10.1007/s00604-018-3204-9
Khodaei R, Ahmady A, Khoshfetrat SM, Kashanian S, Kashanian SM, Omidfar K (2019) Voltammetric immunosensor for E-cadherin promoter DNA methylation using a Fe3O4-citric acid nanocomposite and a screen-printed carbon electrode modified with poly(vinyl alcohol) and reduced graphene oxide. Microchim Acta 186(170):167–173. https://doi.org/10.1007/s00604-019-3234-y
Yin H, Zhou Y, Yang Z, Guo Y, Wang X, Ai S, Zhang X (2015) Electrochemical immunosensor for N6-methyladenosine RNA modification detection. Sensors Actuat B-Chem 221:1–6. https://doi.org/10.1016/j.snb.2015.06.045
Yin H, Wang H, Jiang W, Zhou Y, Ai S (2017) Electrochemical immunosensor for N6-methyladenosine detection in human cell lines based on biotin-streptavidin system and silver-SiO2 signal amplification. Biosens Bioelectron 90:494–500. https://doi.org/10.1016/j.bios.2016.10.066
Dai T, Pu Q, Guo Y, Zuo C, Bai S, Yang Y, Yin D, Li Y, Sheng S, Tao Y, Fang J, Yu W, Xie G (2018) Analogous modified DNA probe and immune competition method-based electrochemical biosensor for RNA modification. Biosens Bioelectron 114:72–77. https://doi.org/10.1016/j.bios.2018.05.018
Pu Q, Li J, Qiu J, Yang X, Li Y, Yin D, Zhang X, Tao Y, Sheng S, Xie G (2017) Universal ratiometric electrochemical biosensing platform based on mesoporous platinum nanocomposite and nicking endonuclease assisted DNA walking strategy. Biosens Bioelectron 94:719–727. https://doi.org/10.1016/j.bios.2017.03.062
Zeng R, Luo Z, Zhang L, Tang D (2018) Platinum Nanozyme-catalyzed gas generation for pressure-based bioassay using Polyaniline nanowires-functionalized Graphene oxide framework. Anal Chem 90(20):12299–12306. https://doi.org/10.1021/acs.analchem.8b03889
Yu Z, Tang Y, Cai G, Ren R, Tang D (2019) Paper electrode-based flexible pressure sensor for point-of-care immunoassay with digital multimeter. Anal Chem 91(2):1222–1226. https://doi.org/10.1021/acs.analchem.8b04635
Kang Y, Murray CB (2010) Synthesis and Electrocatalytic properties of cubic Mn-Pt Nanocrystals (Nanocubes). J Am Chem Soc 132(22):7568–7569. https://doi.org/10.1021/ja100705j
Li Q, Wu L, Wu G, Su D, Lv H, Zhang S, Zhu W, Casimir A, Zhu H, Mendoza-Garcia A, Sun S (2015) New approach to fully ordered fct-FePt nanoparticles for much enhanced Electrocatalysis in acid. Nano Lett 15(4):2468–2473. https://doi.org/10.1021/acs.nanolett.5b00320
Qin Y, Zhang X, Dai X, Sun H, Yang Y, Li X, Shi Q, Gao D, Wang H, Yu NF, Sun SG (2016) Graphene oxide-assisted synthesis of Pt-co alloy Nanocrystals with high-index facets and enhanced Electrocatalytic properties. Small 12(4):524–533. https://doi.org/10.1002/smll.201502669
Sulaiman JE, Zhu S, Xing Z, Chang Q, Shao M (2017) Pt–Ni Octahedra as Electrocatalysts for the ethanol electro-oxidation reaction. ACS Catal 7(8):5134–5141. https://doi.org/10.1021/acscatal.7b01435
Wang H, Yu H, Li Y, Yin S, Xue H, Li X, Xu Y, Wang L (2018) Direct synthesis of bimetallic PtCo mesoporous nanospheres as efficient bifunctional electrocatalysts for both oxygen reduction reaction and methanol oxidation reaction. Nanotechnology 29(17):175403. https://doi.org/10.1088/1361-6528/aaaf3f
Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu PH, Zou L, Jambhekar A, He C, Shi Y (2017) RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543(7646):573–576. https://doi.org/10.1038/nature21671
Polsky R, Gill R, Kaganovsky L, Willner I (2006) Nucleic acid-functionalized Pt nanoparticles: catalytic labels for the amplified electrochemical detection of biomolecules. Anal Chem 78(7):2268–2271. https://doi.org/10.1021/ac0519864
Hurst SJ, Lytton-Jean AK, Mirkin CA (2006) Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem 78(24):8313–8318. https://doi.org/10.1021/ac0613582
Vilhena JG, Dumitru AC, Herruzo ET, Mendieta-Moreno JI, Garcia R, Serena PA, Pérez R (2016) Adsorption orientations and immunological recognition of antibodies on Graphene. Nanoscale 8:13463–13475. https://doi.org/10.1039/C5NR07612A
Wang H, Qi C, He W, Wang M, Jiang W, Yin H, Ai S (2018) A sensitive photoelectrochemical immunoassay of N6–methyladenosine based on dual-signal amplification strategy: Ru doped in SiO2 nanosphere and carboxylated g-C3N4. Biosens Bioelectron 99:281–288. https://doi.org/10.1016/j.bios.2017.07.042
Wang H, Yin H, Huang H, Li K, Zhou Y, Waterhouse GIN, Lin H, Ai S (2018) Dual-signal amplified photoelectrochemical biosensor for detection of N-methyladenosine based on BiVO-110-TiO heterojunction, Ag-mediated cytosine pairs. Biosens Bioelectron 108:89–96. https://doi.org/10.1016/j.bios.2018.02.056
Wang Y, Yin H, Li X, Waterhouse GIN, Ai S (2019) Photoelectrochemical immunosensor for N6-methyladenine detection based on Ru@UiO-66, Bi2O3 and black TiO2. Biosens Bioelectron 131:163–170. https://doi.org/10.1016/j.bios.2019.01.064
Acknowledgments
This research work was financially supported by the National Natural Science Foundation of China (No. 81672112, 81702101), Chongqing Technology Innovation and Application Demonstration Project (cstc2018jscx-msybX0010), Chongqing Special Postdoctoral Science Foundation (Xm2017090), and Top Talent Project of Chongqing Medical University (BJRC201821)
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 799 kb)
Rights and permissions
About this article
Cite this article
Ou, X., Pu, Q., Sheng, S. et al. Electrochemical competitive immunodetection of messenger RNA modified with N6-methyladenosine by using DNA-modified mesoporous PtCo nanospheres. Microchim Acta 187, 31 (2020). https://doi.org/10.1007/s00604-019-4010-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-4010-8