Abstract
Protein p300 is a transcriptional co-activator that participates in many physiological processes including cell cycle control, differentiation and apoptosis. It serves (a) as a protein bridge that links specific transcription factors to the fundamental transcription machinery, (b) as a scaffold to complete multiple transcription cofactors, and (c) as an enzyme for acetylating histone and non-histone proteins. An ultrasensitive electrochemiluminescence (ECL) immunosensor is described here that is based on the use of a magnetic glassy carbon electrode modified with tetrahedral DNA with hollow structure, graphene oxide (GO) and gold nanocrystals. The use of a GO monolayer allows for greater carrying capacity and warrants a wider outer Helmholtz plane. Strong and stable ECL signals were achieved due to antigen-antibody interaction by using the ECL probe Ru(phen)32+. This immunosensor has a response that covers the 0.005 to 80 nM p300 concentration range and has a 1 pM detection limit. It was exploited for the determination of p300 in HeLa cell lysate and (spiked) serum.

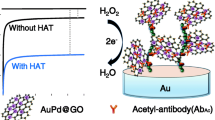
Schematic presentation of an ultrasensitive Faraday-cage electrochemiluminescence immunosensor toward the transcriptional co-activator p300 analysis is presented based on a graphene oxide monolayer and tetrahedral DNA-mediated signal amplification.





Similar content being viewed by others
References
Michaelides MR, Kluge A, Patane M, Van Drie JH, Wang C, Hansen TM, Risi RM, Mantei R, Hertel C, Karukurichi K, Nesterov A, McElligott D, Vries P, Langston JW, Cole PA, Marmorstein R, Liu H, Lasko L, Bromberg KD, Lai A, Kesicki EA (2018) Discovery of spiro oxazolidinediones as selective, orally bioavailable inhibitors of p300/CBP histone acetyltransferases. ACS Med Chem Lett 9:28–33
Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 28:707–719
Biel M, Wascholowski V, Giannis A (2005) Epigenetics-an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed 44:3186–3216
Dekker FJ, Haisma HJ (2009) Histone acetyl transferases as emerging drug targets. Drug Discov Today 14:942–948
Selvi RB, Kundu TK (2010) Reversible acetylation of chromatin: implication in regulation of gene expression, disease and therapeutics. Biotechnol J 4:375–390
Kamei Y, Xu L, Heinzel T, Torchia J, Rosenfeld MG (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403–414
Goodman RH, Smolik S (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev 14:1553–1577
Nakajima T, Fukamizu A, Takahashi J, Gage FH, Fisher T, Blenis J, Montminy MR (1996) The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell 86:465–474
Chan HM, Thangue NBL (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114:2363–2373
Kim TK, Kim TH, Maniatis T (1998) Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon- enhanceosome in vitro. Proc Natl Acad Sci U S A 95:12191–12196
Miyamoto S, Kawamura T, Morimoto T, Ono K, Wada H, Kawase Y, Matsumori A, Nishio R, Kita T, Hasegawa K (2006) Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation 113:679–690
Ait-Si-Ali S, Ramirez S, Robin P, Trouche D, Harel-Bellan A (1998) A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res 26:3869–3870
Poveda A, Sendra R (2008) An easy assay for histone acetyltransferase activity using a phosphor imager. Anal Biochem 383:296–300
Chen SY, Li Y, Hu YF, Han YT, Huang Y, Nie Z, Yao SZ (2015) Nucleic acid-mimicking coordination polymer for label-free fluorescent activity assay of histone acetyltransferases. Chem Commun 51:4469–4472
Hu YF, Chen SY, Han YT, Chen HJ, Wang Q, Nie Z, Huang Y, Yao SZ (2015) Unique electrocatalytic activity of a nucleic acid-mimicking coordination polymer for the sensitive detection of coenzyme A and histone acetyltransferase activity. Chem Commun 51:17611–17614
Han YT, Li P, Xu YT, Li H, Song ZL, Nie Z, Zhuo C, Yao SZ (2015) Fluorescent nanosensor for probing histone acetyltransferase activity based on acetylation protection and magnetic graphitic nanocapsules. Small 11:877–885
Zou Y, Wang ZH, Zhang HX, Liu Y (2018) A novel electrogenerated chemiluminescence biosensor for histone acetyltransferases activity analysis and inhibition based on mimetic superoxide dismutase of tannic acid assembled nanoprobes. Biosens Bioelectron 122:205–210
Rasheed PA, Lee JS (2017) Recent advances in optical detection of dopamine using nanomaterials. Microchim Acta 184:1239–1266
Richter MM (2004) Electrochemiluminescence (ECL). Chem Rev 104:3003–3036
Voci S, Goudeau B, Valenti G, Lesch A, Jović M, Rapino S, Paolucci F, Arbault S, Sojic N (2018) Surface-confined electrochemiluminescence microscopy of cell membranes. J Am Chem Soc 140:14753–14760
Zhang Y, Lu F, Yan ZQ, Wu D, Ma HM, Du B, Wei Q (2015) Electrochemiluminescence immunosensing strategy based on the use of Au@Ag nanorods as a peroxidase mimic and NH4CoPO4 as a supercapacitive supporter: application to the determination of carcinoembryonic antigen. Microchim Acta 182:1421–1429
Chen ZH, Liu Y, Wang YZ, Zhao X, Li JH (2013) Dynamic evaluation of cell surface n-glycan expression via an electrogenerated chemiluminescence biosensor based on concanavalin a-integrating gold-nanoparticle-modified Ru(bpy)3 2+-doped silica nanoprobe. Anal Chem 85:4431–4438
Guo YS, Shang XX, Liu F, Hu YH, Li S, Liu J, Wu F (2018) Novel enhancer for luminol-AuNP electrochemiluminescence and decoration on RNA membranes for effective cytosensing. ACS Appl Bio Mater 1:1647–1655
Yang SS, Jiang MH, Chai YQ, Yuan R, Zhuo Y (2018) Application of antibody-powered triplex-DNA nanomachine to electrochemiluminescence biosensor for the detection of anti-digoxigenin with improved sensitivity versus cycling strand displacement reaction. ACS Appl Mater Interfaces 10:38648–38655
Guo ZY, Sha YH, Hu YF, Wang S (2016) In-electrode vs. on-electrode: ultrasensitive faraday cage-type electrochemiluminescence immunoassay. Chem Commun 52:4621–4624
Lu J, Wu L, Hu YF, Wang S, Guo ZY (2018) Ultrasensitive faraday cage-type electrochemiluminescence assay for femtomolar miRNA-141 via graphene oxide and hybridization chain reaction-assisted cascade amplification. Biosens Bioelectron 109:13–19
Xu JH, Wang YZ, Hu SS (2017) Nanocomposites of graphene and graphene oxides: synthesis, molecular functionalization and application in electrochemical sensors and biosensors. A review. Microchim Acta 184:1–44
Lin L, Deng B, Sun JY, Peng HL, Liu ZF (2018) Bridging the gap between reality and ideal in chemical vapor deposition growth of graphene. Chem Rev 118:9281–9343
Wang Y, Tang LH, Li ZH, Lin YH, Li JH (2014) In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells. Nat Protoc 9:1944–1955
Wang Y, Lu J, Tang LH, Chang HX, Li JH (2009) Graphene oxide amplified electrogenerated chemiluminescence of quantum dots and its selective sensing for glutathione from thiol-containing compounds. Anal Chem 81:9710–9715
Wang Y, Li ZH, Hu DH, Lin CT, Li JH, Lin YH (2010) Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc 132:9274–9276
Chen D, Feng HB, Li JH (2012) Graphene oxide: preparation, functionalization, and electrochemical applications. Chem Rev 112:6027–6053
Tang LH, Chang HX, Liu Y, Li JH (2012) Duplex DNA/graphene oxide biointerface: from fundamental understanding to specific enzymatic effects. Adv Funct Mater 22:3083–3088
Tang LH, Wang Y, Li JH (2015) The graphene/nucleic acid nanobiointerface. Chem Soc Rev 44:6954–6980
Liu Y, Lin CX, Li HY, Yan H (2005) Aptamer-directed self-assembly of protein arrays on a DNA nanostructure. Angew Chem 117:4407–4412
Huang YL, Mo S, Gao ZF, Chen JR, Lei JL, Luo HQ, Li NB (2017) Amperometric biosensor for microRNA based on the use of tetrahedral DNA nanostructure probes and guanine nanowire amplification. Microchim Acta 184:2597–2604
Mitchell N, Schlapak R, Kastner M, Armitage D, Chrzanowski W, Riener J, Hinterdorfer P, Ebner A, Howorka S (2009) A DNA nanostructure for the functional assembly of chemical groups with tunable stoichiometry and defined nanoscale geometry. Angew Chem Int Ed 48:525–527
Giovanni M, Setyawati MI, Tay CY, Qian H, Kuan WS, Leong DT (2015) Electrochemical quantification of escherichia coli with DNA nanostructure. Adv Funct Mater 25:3840–3846
Yuan L, Giovanni M, Xie JP, Fan CH, Leong DT (2014) Ultrasensitive IgG quantification using DNA nano-pyramids. NPG Asia Mater 6:e112
Acknowledgements
Financial support from the National Natural Science Foundation of China (21605089 and 81773483), the Ningbo Municipal Natural Science Foundation (2017A610228 and 2018A610217), the Open Subject of State Key Laboratory of Chemo/Biosensing and Chemometrics (2016001), and Zhejiang Provincial Natural Science Foundation of China (LY13B070013) are gratefully acknowledged. This work was also sponsored by K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 619 kb)
Rights and permissions
About this article
Cite this article
Hu, Y., Zhang, Q., Hu, D. et al. Ultrasensitive electrochemiluminescence immunosensor for the transcriptional co-activator p300 by using a graphene oxide monolayer and tetrahedral DNA-mediated signal amplification. Microchim Acta 186, 325 (2019). https://doi.org/10.1007/s00604-019-3435-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3435-4