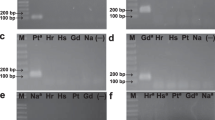
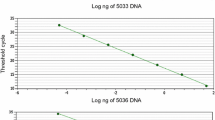
Abstract
Rhizophagus irregularis (previously named Glomus irregulare) is one of the most widespread and common arbuscular mycorrhizal fungal (AMF) species. It has been recovered worldwide in agricultural and natural soils, and the isolate DAOM-197198 has been utilized as a commercial inoculant for two decades. Despite the ecological and economical importance of this taxon, specific markers for quantification of propagules by quantitative real-time PCR (qPCR) are extremely limited and none have been rigorously validated for quality control of manufactured products such as biofertilizers. From the sequencing of 14 complete AMF mitochondrial (mt) genomes, a qPCR assay using a hydrolysis probe designed in the single copy cox3-rnl intergenic region was tested and validated to specifically and accurately quantify the spores of R. irregularis isolate DAOM-197198. Specificity tests were performed using standard PCR and qPCR, and results clearly showed that the primers specifically amplified the isolate DAOM-197198, yielding a PCR product of 106 bp. According to the qPCR analyses on spores produced in vitro, the average copy number of mt genomes per spore was 3172 ± 304 SE (n = 6). Quantification assays were successfully undertaken on known and unknown samples in liquid suspensions and commercial dry formulations to show the accuracy, precision, robustness, and reproducibility of the qPCR assay. This study provides a powerful molecular toolkit specifically designed to quantify spores of the model AMF isolate DAOM-197198. The approach of molecular toolkit used in our study could be applied to other AMF taxa and will be useful to research institutions and governmental and industrial laboratories running routine quality control of AMF-based products.



Similar content being viewed by others
References
Alkan N, Gadkar V, Coburn J, Yarden O, Kapulnik Y (2004) Quantification of the arbuscular mycorrhizal fungus Glomus intraradices in host tissue using real-time polymerse chain reaction. New Phytol 161:877–885
Alkan N, Gadkar V, Yarden O, Kapulnik Y (2006) Analysis of quantitative interactions between two species of arbuscular mycorrhizal fungi, Glomus mosseae and G. intraradices, by real-time PCR. Appl Environ Microbiola 72:4192–4199
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Beaudet D, de IE l P, Labridy M et al (2015) Intraisolate mitochondrial genetic polymorphism and gene variants coexpression in arbuscular mycorrhizal fungi. Genome Biol Evol 7:218–227
Beaudet D, Nadimi M, Hijri M (2013a) Rapid mitochondrial genome evolution through invasion of mobile elements in two closely related species of arbuscular mycorrhizal fungi. PLoS One 8:e60768
Beaudet D, Terrat Y, Halary S, de la Providencia IE, Hijri M (2013b) Mitochondrial genome rearrangements in Glomus species triggered by homologous recombination between distinct mtDNA haplotypes. Genome Biol Evol 5:1628–1643
Boon E, Zimmerman E, Lang BF, Hijri M (2010) Intra-isolate genome variation in arbuscular mycorrhizal fungi persists in the transcriptome. J Evol Biol 23:1519–1527
Center for Drug Evaluation and Research (2001) Guidance for Industry Bioanalytical Method Validation. http://www.fda.gov/downloads/Drugs/…/Guidances/ucm070107.pdf. Accessed 22 December 2015.
Clapp J, Fitter AH, Young JP (1999) Ribosomal small subunit sequence variation within spores of an arbuscular mycorrhizal fungus, Scutellospora sp. Mol Ecol 8:915–921
Clapp J, Rodriguez A, Dodd C (2001) Inter- and intra-isolate rRNA large subunit variation in Glomus coronatum spores. New Phytol 149:539–554
Corradi N, Croll D, Colard A, Kuhn G, Ehinger M, Sanders IR (2007) Gene copy number polymorphisms in an arbuscular mycorrhizal fungal population. Appl Environ Microbiol 73:366–369
Couillerot O, Ramirez-Trujillo A, Walker V, von Felten A, Jansa J, Maurhofer M, Défago G, Prigent-Combaret C, Comte G, Caballero-Mellado J, Moënne-Loccoz Y (2013) Comparison of prominent Azospirillum strains in Azospirillum-Pseudomonas-Glomus consortia for promotion of maize growth. Appl Microbiol Biotechnol 97:4639–4649
Daubois L, Beaudet D, Hijri M, de la Providencia IE (2016) Independent mitochondrial and nuclear exchanges arising in Rhizophagus irregularis crossed-isolates support the presence of a mitochondrial segregation mechanism. BMC Microbiol 16(11)
das Neves RP, Jones NS, Andreu L, Gupta R, Enver T et al (2010) Connecting variability in global transcription rate to mitochondrial variability. PLoS Biol 8(12):e1000560
de la Providencia IE, Nadimi M, Beaudet D, Morales GR, Hijri M (2013) Detection of a transient mitochondrial DNA heteroplasmy in the progeny of crossed genetically divergent isolates of arbuscular mycorrhizal fungi. New Phytol 200:211–221
Filion M, St-Arnaud M, Jabaji-Hare SH (2003) Direct quantification of fungal DNA from soil substrate using real-time PCR. J Microbiol Methods 53:67–76
Formey D, Molès M, Haouy A, Savelli B, Bouchez O, Becard G, Roux C (2012) Comparative analysis of mitochondrial genomes of Rhizophagus irregularis—syn. Glomus irregulare—reveals a polymorphism induced by variability generating elements. New Phytol 196:1217–1227
Fortin JA, Plenchette C, Piché Y (2015) Les mycorhizes, Multimondesth edn
Gamper HA, Young JPW, Jones DL, Hodge A (2008) Real-time PCR and microscopy: are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance? Fungal Genet Biol 45:581–596
Gianinazzi S, Gollotte A, Binet M-N et al (2010) Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20:519–530
Giovannetti M, Azzolini D, Citernesi AS (1999) Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl Environ Microbiol 65:5571–5575
Hausner G (2003) Fungal mitochondrial genomes, plasmids and introns. Appl Environ Microbiol 3:101–131
Hijri M, Niculita H, Sanders IR (2007) Molecular characterization of chromosome termini of the arbuscular mycorrhizal fungus Glomus intraradices (Glomeromycota). Fungal Genet Biol 44:1380–1386
Hijri M, Sanders IR (2005) Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature 433:160–163
Hijri M (2016) Analysis of a large dataset form field mycorrhizal inoculation trials on potato showed highly significant increase in yield. Mycorrhiza 26(3):209–214
International Conference on Harmonization guideline (1995) Structure and content of clinical study Reports E3. http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Efficacy/E3/E3_Guideline.pdf
International Conference on Harmonization (ICH). Technical requirements for registration of pharmaceuticals for human use. Topic Q2 (R1): Validation of analytical procedures: text and methodology. Geneva, Switzerland, 2005. http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q2_R1/Step4/Q2_R1__Guideline.pdf Accessed 22 December 2015.
Isayenkov S, Fester T, Hause B (2004) Rapid determination of fungal colonization and arbuscule formation in roots of Medicago truncatula using real-time (RT) PCR. J Plant Physiol 161:1379–1383
Jansa J, Smith FA, Smith SE (2008) Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol 177:779–789
Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, Palmer TM, West SA, Vandenkoornhuyse P, Jansa J, Bucking H (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333:880–882
Koide RT, Mosse B (2004) A history of research on arbuscular mycorrhiza. Mycorrhiza 14:145–163
Krak K, Janouskova M, Caklova P, Vosatka M, Storchova H (2012) Intraradical dynamics of two coexisting isolates of the arbuscular mycorrhizal fungus Glomus intraradices sensu lato as estimated by real-time PCR of mitochondrial DNA. Appl Environ Microbiol 78(10):3630–3637
Lang BF, Hijri M (2009) The complete Glomus intraradices mitochondrial genome sequence—a milestone in mycorrhizal research. New Phytol 183:3–6
Lee J, Young JPW (2009) The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol 183:200–211
Lin K, Limpens E, Zhang Z et al (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10:e1004078
Marleau J, Dalpé Y, St-Arnaud M, Hijri M (2011) Spore development and nuclear inheritance in arbuscular mycorrhizal fungi. BMC Evol Biol 11:51
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65(1–2):55–63
Nadimi M, Stefani FOP, Hijri M (2015) The mitochondrial genome of the glomeromycete Rhizophagus sp. DAOM 213198 reveals an unusual organization consisting of two circular chromosomes. Genome Biol Evol 7(1):96–105
Nadimi M, Daubois L, Hijri M (2016) Mitochondrial comparative genomics and phylogenetic signal assessment of mtDNA among arbuscular mycorrhizal fungi. Mol Phylogenet Evol. doi:10.1016/j.ympev.2016.01.009, In Press
Nadimi M, Beaudet D, Forget L, Hijri M, Lang BF (2012) Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol Biol Evol 29(9):2199–2210
Peyret-Guzzon M, Stockinger H, Bouffaud M-L, Farcy P, Wipf D, Redecker D (2016) Arbuscular mycorrhizal fungal communities and Rhizophagus irregularis populations shift in response to short-term ploughing and fertilisation in a buffer strip. Mycorrhiza 26:33–46
Podeszfinski C, Dalpé Y, Charest C (2002) In situ turfgrass establishment: I. Responses to arbuscular mycorrhizae and fertilization. J Sustain Agric 20:57–74
Raab PA, Breenwald A, Redecker D (2005) Mitochondrial large ribosomal subunit sequences are homogeneous within isolates of Glomus (arbuscular mycorrhizal fungi, Glomeromycota). Mycol Res 109:1315–1322
Redecker D, Hijri M, Dulieu H, Sanders IR (1999) Phylogenetic analysis of a dataset of fungal 5.8S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet Biol 28:238–244
Sanders IR, Alt M, Groppe K, Boller T, Wiemken A (1995) Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol 130:419–427
Shrivastava A, Gupta VB (2011) Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci 2:21–25
Smith SE, Read DJ (2010) Mycorrhizal Symbiosis. Academic, Cambridge
Takada-Hoshino Y, Matsumoto N (2004) An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ 19:13–19
Thonar C, Erb A, Jansa J (2012) Real-time PCR to quantify composition of arbuscular mycorrhizal fungal communities—marker design, verification, calibration and field validation. Mol Ecol Resour 12:219–232
Tisserant E, Malbreil M, Kuo A et al (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci 110:20117–20122
Acknowledgements
This work is a part of a research project organized and coordinated by Premier Tech. The authors are grateful for financial support from NSERC Cooperative Research and Development (grant number CRDJP 468828-14), Premier Tech, and CRIBIQ. We would like to thank Dr. Serge Gagné, Dr. Younes Machrafi, Nicolas Bertrand, Éric Dion, François Gobeil, and Dr Alain Bélanger for recommendations and comments. We also thank Dr. David Morse and Dr. Karen Fisher-Favret for English editing and Dr D. Janos and two anonymous reviewers for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Amine Badri and Franck O. P. Stefani contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Esm 1
(DOCX 111 kb)
Rights and permissions
About this article
Cite this article
Badri, A., Stefani, F.O.P., Lachance, G. et al. Molecular diagnostic toolkit for Rhizophagus irregularis isolate DAOM-197198 using quantitative PCR assay targeting the mitochondrial genome. Mycorrhiza 26, 721–733 (2016). https://doi.org/10.1007/s00572-016-0708-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-016-0708-1