Abstract
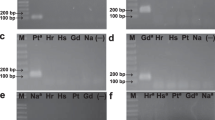
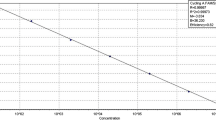
A real-time quantitative TaqMan-PCR was established for the absolute quantification of extramatrical hyphal biomass of the ectomycorrhizal fungus Piloderma croceum in pure cultures as well as in rhizotron samples with non-sterile peat substrate. After cloning and sequencing of internal transcribed spacer (ITS) sequences ITS1/ITS2 and the 5.8S rRNA gene from several fungi, including Tomentellopsis submollis, Paxillus involutus, and Cortinarius obtusus, species-specific primers and a dual-labelled fluorogenic probe were designed for Piloderma croceum. The dynamic range of the TaqMan assay spans seven orders of magnitude, producing an online-detectable fluorescence signal during the cycling run that is directly related to the starting number of ITS copies present. To test the confidence of the PCR-based quantification results, the hyphal length of Piloderma croceum was counted under the microscope to determine the recovery from two defined but different amounts of agar-cultivated mycelia. Inspection of the registered Ct values (defined as that cycle number at which a statistically significant increase in the reporter fluorescence can first be detected) in a 10-fold dilution series of template DNA represents a suitable and stringent quality control standard for exclusion of false PCR-based quantification results. The fast real-time PCR approach enables high throughput of samples, making this method well suited for quantitative analysis of ectomycorrhizal fungi in communities of natural and artificial ecosystems, so long as applicable DNA extraction protocols exist for different types of soil.


Similar content being viewed by others
References
Agerer R (ed) (1987–1998) Colour atlas of ectomycorrhizae. 1st–12th edn. Einhorn, Schwäbisch Gmünd, Germany
Agerer R (1990) Studies on ectomycorrhizae. XXIV. Ectomycorrhizae of Chroogomphus helveticus and C. rutilus (Gomphidiaceae, Basidiomycetes) and their relationship to those of Suillus and Rhizopogon. Nova Hedwigia 50:1–63
Agerer R (1991) Studies on ectomycorrhizae. XXXIV. Mycorrhizae of Gomphidius glutinosus and of G. roseus with some remarks on Gomphidiaceae (Basidiomycetes). Nova Hedwigia 53:127–170
Agerer R (2001) Exploration types of ectomycorrhizal mycelial systems. A proposal to classify mycorrhizal mycelial systems with respect to their ecologically important contact area with the substrate. Mycorrhiza 11:107–114
Agerer R, Rambold G (1998) DEEMY, a DELTA-based information system for characterization and determination of ectomycorrhizae. Version 1.1. Institute for Systematic Botany, Section Mycology, University of Munich
Agerer R, Müller WR, Bahnweg G (1996) Ectomycorrhizae of Rhizopogon subcaerulescens on Tsuga heterophylla. Nova Hedwigia 63:397–416
Allen MF (1991) The ecology of mycorrhiza. Cambridge University Press, Cambridge
Bach HJ, Tomanova J, Schloter M, Munch JC (2002) Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J Microbiol Methods 49:235–245
Bahnweg G, Schulze S, Möller EM, Rosenbrock H, Langebartels C, Sandermann H (1998) DNA isolation from recalcitrant materials such as tree roots, bark, and forest soil for the detection of fungal pathogens by polymerase chain reaction. Anal Biochem 262:79–82
Bååth E, Söderström B (1979) Fungal biomass and fungal immobilization of plant nutrients in Swedish coniferous forest soils. Rev Ecol Biol Sol 16:477–489
Böhm J, Hahn A, Schubert R, Bahnweg G, Adler N, Nechwatal J, Oehlmann R, Oßwald W (1999) Real-time quantitative PCR: DNA determination in isolated spores of the mycorrhizal fungus Glomus mosseae and monitoring of Phytophthora infestans and Phytophthora citricola in their respective host plants. J Phytopathol 147:409–416
Brand F, Agerer R (1988) Studien an Ektomykorrhizen. XIII. Drei häufige Ektomykorrhizen der Buche (Fagus sylvatica L.). Charakterisierung und unsterile Kultivierung von Buchenektomykorrhizen. Sydowia 40:1–37
Buscot F, Munch JC, Charcosset JY, Gardes M, Nehls U, Hampp R (2000) Recent advances in exploring physiology and biodiversity of ectomycorrhizas highlight the functioning of these symbioses in ecosystems. FEMS Microbiol Rev 24:601–614
Cullen DW, Hirsch PR (1998) Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol Biochem 30:983–993
Desjardin LE, Chen Y, Perkins MD, Teixeira L, Cave MD, Eisenach KD (1998) Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J Clin Microbiol 36:1964–1968
Fogel R, Hunt G (1978) Fungal and arboreal biomass in a western Oregon Douglas-fir ecosystem: distribution patterns and turnover. Can J For Res 9:245–256
Frankland JC, Lindley DK, Swift MJ (1978) A comparison of two methods for the estimation of mycelial biomass in leaf litter. Soil Biol Biochem 10:323–333
Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994
Higgins DG, Sharp PM (1988) CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene 73:237–244
Horton TR, Bruns TD (2001) The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol Ecol 10:1855–1871
Ineichen K, Wiemken V, Wiemken A (1995) Shoots, roots and ectomycorrhiza formation of pine seedlings at elevated atmospheric carbon dioxide. Plant Cell Environ 18:703–707
Kowalchuk GA (1999) New perspectives towards analysing fungal communities in terrestrial environments. Curr Opin Biotechnol 10:247–51
Kunzweiler K, Kottke I (1986) Quantifizierung von Myzel im Waldboden. In: Einsele G (ed) Das landschaftsökologische Forschungsprojekt Naturpark Schönbuch. VCH , Weinheim, Germany, pp 429–441
Larsen MJ, Smith JE, McKay D (1997) On Piloderma bicolor and the closely related P. byssinum, Piloderma croceum and P. fallax. Mycotaxon 63:1–8
Manzin A, Solforosi L, Bianchi D, Gabrielli A, Giostra F, Bruno S, Clementi M (1995) Viral load in samples from hepatitis C virus (HCV)-infected patients with various clinical conditions. Res Virol 146:279–284
Marx DH (1969) The influence of ectotrophic mycorrhizal fungi on the resistance of pine roots to pathogenic infections. I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 59:153–163
Matyssek R, Schnyder H, Elstner EF, Munch JC, Pretzsch H, Sandermann H (2002) Growth and parasite defence in plants: the balance between resource sequestration and retention: in lieu of a guest editorial. Plant Biol 4:133–136
Mensink E, van de Locht A, Schattenberg A, Linders E, Schaap N, Geurts van Kessel A, De Witte T (1998) Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukaemia patients using real-time quantitative RT-PCR. Br J Haematol 102:768–774
Nazar RN, Robb EJ, Hu X, Volossiouk T, Lee SW (1997) Development of PCR-based diagnostics for soil borne plant pathogens. J Plant Biol 40:176–81
Needleman SB, Wunsch CD (1970) Description of the method used in PCOMPARE and SCANISM. J Mol Biol 48:443–453
Peterson RL, Chakravarty P (1991) Techniques in synthesizing ectomycorrhiza. In: Norris JR, Read DJ, Varma AK (eds) Techniques for the study of mycorrhiza. Methods Microbiol 23:75–106
Raidl S (1997) Studien zur Ontogenie an Rhizomorphen von Ektomykorrhizen. Bibl Mycol 169:1–184
Read DJ (1992) The mycorrhizal mycelium. In: Allen MF (ed) Mycorrhizal functioning. An integrative plant–fungal process. Chapman & Hall, New York, pp 102–133
Sanseverino J, Layton AC, Sayler GS (2002) Detection of polychlorinated biphenyl-degrading organisms in soil. In: Aquino de Muro M, Rapley R (eds) Gene probes: principles and protocols. Methods in Molecular Biology, vol 179. Humana, Totowa, N. J., pp 193–209
Schild TA (1996) Einführung in die Real-Time TaqManTM PCR-Technologie. Applied Biosystems GmbH, Weiterstadt
Schubert R, Bahnweg G, Nechwatal J, Jung T, Cooke DEL, Duncan JM, Müller-Starck G, Langebartels C, Sandermann H, Oßwald W (1999) Detection and quantification of Phytophthora species which are associated with root-rot diseases in European deciduous forests by species-specific polymerase chain reaction. Eur J For Pathol 29:169–188
Smith SE, Read DJ (eds) (1997) Mycorrhizal symbiosis. Academic, San Diego
Tan S-L, Dossett M, Katze MG (1998) Extraction of genomic DNA suitable for PCR analysis from dried plant rhizomes/roots. BioTechniques 25:796–801
Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A (eds) (1999) Methods of soil analysis. Part II. Microbiological and biochemical properties. Soil Science Society of America, Madison, Wis
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Snisky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–321
Acknowledgement
The study was supported by the Deutsche Forschungsgemeinschaft (SFB 607, A1/A9/B7).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schubert, R., Raidl, S., Funk, R. et al. Quantitative detection of agar-cultivated and rhizotron-grown Piloderma croceum Erikss. & Hjortst. by ITS1-based fluorescent PCR. Mycorrhiza 13, 159–165 (2003). https://doi.org/10.1007/s00572-002-0212-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-002-0212-7