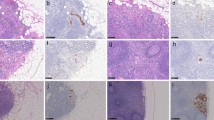
Abstract
Background
When endoscopically resected specimens of early colorectal cancer (CRC) show high-risk features, surgery should be performed based on current guidelines because of the high-risk of lymph node metastasis (LNM). The aim of this study was to determine the utility of an artificial intelligence (AI) with deep learning (DL) of hematoxylin and eosin (H&E)-stained endoscopic resection specimens without manual-pixel-level annotation for predicting LNM in T1 CRC. In addition, we assessed AI performance for patients with only submucosal (SM) invasion depth of 1000 to 2000 μm known to be difficult to predict LNM in clinical practice.
Methods
H&E-stained whole slide images (WSIs) were scanned for endoscopic resection specimens of 400 patients who underwent endoscopic treatment for newly diagnosed T1 CRC with additional surgery. The area under the curve (AUC) of the receiver operating characteristic curve was used to determine the accuracy of AI for predicting LNM with a fivefold cross-validation in the training set and in a held-out test set.
Results
We developed an AI model using a two-step attention-based DL approach without clinical features (AUC, 0.764). Incorporating clinical features into the model did not improve its prediction accuracy for LNM. Our model reduced unnecessary additional surgery by 15.1% more than using the current guidelines (67.4% vs. 82.5%). In patients with SM invasion depth of 1000 to 2000 μm, the AI avoided 16.1% of unnecessary additional surgery than using the JSCCR guidelines.
Conclusions
Our study is the first to show that AI trained with DL of H&E-stained WSIs has the potential to predict LNM in T1 CRC using only endoscopically resected specimens with conventional histologic risk factors.




Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- AI:
-
Artificial intelligence
- AM:
-
Attention module
- AS:
-
Attention score
- CRC:
-
Colorectal cancer
- CV:
-
Cross-validation
- DCNN:
-
Deep convolution neural network
- DL:
-
Deep learning
- EMR:
-
Endoscopic mucosal resection
- ESD:
-
Endoscopic submucosal dissection
- FE:
-
Feature extractor
- FV:
-
Feature vector
- H&E:
-
Hematoxylin and eosin
- IQR:
-
Interquartile range
- JSCCR:
-
Japanese Society for Cancer of the Colon and Rectum
- LNM:
-
Lymph node metastasis
- LVI:
-
Lymphovascular invasion
- RF:
-
Random forest
- ROI:
-
Regions of interest
- SM:
-
Submucosal
- ROC:
-
The receiver operating characteristic
- WSIs:
-
Whole slide images
References
Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–53. https://doi.org/10.1002/ijc.31937.
Wong MCS, Ding H, Wang J, et al. Prevalence and risk factors of colorectal cancer in Asia. Intest Res. 2019;17(3):317–29. https://doi.org/10.5217/ir.2019.00021.
Fujimori T, Kawamata H, Kashida H. Precancerous lesions of the colorectum. J Gastroenterol. 2001;36(9):587–94. https://doi.org/10.1007/s005350170041.
Morson BC, Whiteway JE, Jones EA, et al. Histopathology and prognosis of malignant colorectal polyps treated by endoscopic polypectomy. Gut. 1984;25(5):437–44. https://doi.org/10.1136/gut.25.5.437(ineng).
Minamoto T, Mai M, Ogino T, et al. Early invasive colorectal carcinomas metastatic to the lymph node with attention to their nonpolypoid development. Am J Gastroenterol. 1993;88(7):1035–9 ((in eng)).
Kitajima K, Fujimori T, Fujii S, et al. Correlations between lymph node metastasis and depth of submucosal invasion in submucosal invasive colorectal carcinoma: a Japanese collaborative study. J Gastroenterol. 2004;39(6):534–43.
Kyzer S, Begin LR, Gordon PH, et al. The care of patients with colorectal polyps that contain invasive adenocarcinoma Endoscopic polypectomy or colectomy? Cancer. 1992;70(8):2044–50.
Watanabe T, Muro K, Ajioka Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clini Oncol. 2018;23(1):1–34. https://doi.org/10.1007/s10147-017-1101-6.
Ramirez M, Schierling S, Papaconstantinou HT, et al. Management of the malignant polyp. Clin Colon Rectal Surg. 2008;21(4):286–90. https://doi.org/10.1055/s-0028-1089944(ineng).
Aarons CB, Shanmugan S, Bleier JI. Management of malignant colon polyps: current status and controversies. World J Gastroenterol. 2014;20(43):16178–83. https://doi.org/10.3748/wjg.v20.i43.16178(ineng).
Pérez I, Fernández L, Sánchez-Ramón S, et al. Reliability evaluation of four different assays for therapeutic drug monitoring of infliximab levels. Therap Adv Gastroenterol. 2018;11:1756284818783613–1756284818783613. https://doi.org/10.1177/1756284818783613(ineng).
Nivatvongs S, Rojanasakul A, Reiman HM, et al. The risk of lymph node metastasis in colorectal polyps with invasive adenocarcinoma. Dis Colon Rectum. 1991;34(4):323–8. https://doi.org/10.1007/bf02050592(ineng).
Netzer P, Forster C, Biral R, et al. Risk factor assessment of endoscopically removed malignant colorectal polyps. Gut. 1998;43(5):669–74. https://doi.org/10.1136/gut.43.5.669(ineng).
Cooper HS. Surgical pathology of endoscopically removed malignant polyps of the colon and rectum. Am J Surg Pathol. 1983;7(7):613–23.
Coverlizza S, Risio M, Ferrari A, et al. Colorectal adenomas containing invasive carcinoma. Pathologic assessment of lymph node metastatic potential. Cancer. 1989;64(9):1937–47.
Colacchio TA, Forde KA, Scantlebury V. Endoscopic polypectomy: inadequate treatment for invasive colorectal carcinoma. Ann Surg. 1982;194:704–7.
Choi YS, Kim WS, Hwang SW, et al. Clinical outcomes of submucosal colorectal cancer diagnosed after endoscopic resection: a focus on the need for surgery. Intest Res. 2020;18(1):96–106. https://doi.org/10.5217/ir.2019.00092.
Kojima M, Puppa G, Kirsch R, et al. Blood and lymphatic vessel invasion in pT1 colorectal cancer: an international concordance study. J Clin Pathol. 2015;68(8):628–32. https://doi.org/10.1136/jclinpath-2014-202805(ineng).
Kouyama Y, Kudo SE, Miyachi H, et al. Practical problems of measuring depth of submucosal invasion in T1 colorectal carcinomas. Int J Colorectal Dis. 2016;31(1):137–46. https://doi.org/10.1007/s00384-015-2403-7(ineng).
Barel F, Auffret A, Cariou M, et al. High reproducibility is attainable in assessing histoprognostic parameters of pT1 colorectal cancer using routine histopathology slides and immunohistochemistry analyses. Pathology. 2019;51(1):46–54. https://doi.org/10.1016/j.pathol.2018.10.007(ineng).
Ichimasa K, Kudo SE, Miyachi H, et al. Current problems and perspectives of pathological risk factors for lymph node metastasis in T1 colorectal cancer: systematic review. Dig Endosc. 2021. https://doi.org/10.1111/den.14220(ineng).
Norgeot B, Glicksberg BS, Butte AJ. A call for deep-learning healthcare. Nat Med. 2019;25(1):14–5. https://doi.org/10.1038/s41591-018-0320-3.
Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci. 2018;115(13):E2970–9. https://doi.org/10.1073/pnas.1717139115.
Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. npj Digit Med. 2019;2(1):48. https://doi.org/10.1038/s41746-019-0112-2.
Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–67. https://doi.org/10.1038/s41591-018-0177-5.
Schmauch B, Romagnoni A, Pronier E, et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. 2020;11(1):3877. https://doi.org/10.1038/s41467-020-17678-4.
Ichimasa K, Kudo SE, Mori Y, et al. Artificial intelligence may help in predicting the need for additional surgery after endoscopic resection of T1 colorectal cancer. Endoscopy. 2018;50(3):230–40. https://doi.org/10.1055/s-0043-122385(ineng).
Takamatsu M, Yamamoto N, Kawachi H, et al. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Progr Biomed. 2019;178:155–61. https://doi.org/10.1016/j.cmpb.2019.06.022.
Kudo S-e, Ichimasa K, Villard B, et al. Artificial intelligence system to determine risk of T1 colorectal cancer metastasis to lymph node. Gastroenterology. 2001. https://doi.org/10.1053/j.gastro.2020.09.027.
Egashira Y, Yoshida T, Hirata I, et al. Analysis of pathological risk factors for lymph node metastasis of submucosal invasive colon cancer. Mod Pathol. 2004;17(5):503–11. https://doi.org/10.1038/modpathol.3800030.
Tanaka S, Haruma K, Oh-E H, et al. Conditions of curability after endoscopic resection for colorectal carcinoma with submucosally massive invasion. Oncol Rep. 2000;7(4):783–91. https://doi.org/10.3892/or.7.4.783.
Nakadoi K, Tanaka S, Kanao H, et al. Management of T1 colorectal carcinoma with special reference to criteria for curative endoscopic resection. J Gastroenterol Hepatol. 2012. https://doi.org/10.1111/j.1440-1746.2011.07041.x(ineng).
Vermeer NCA, Backes Y, Snijders HS, et al. National cohort study on postoperative risks after surgery for submucosal invasive colorectal cancer. BJS Open. 2018;3(2):210–7. https://doi.org/10.1002/bjs5.50125(ineng).
Ilse M, Tomczak J, Welling M, editors. Attention-based deep multiple instance learning. International conference on machine learning. 2018 PMLR
Lu MY, Zhao M, Shady M, et al. Deep learning-based computational pathology predicts origins for cancers of unknown primary. arXiv preprint arXiv:200613932. 2020.
Echle A, Rindtorff NT, Brinker TJ, et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer. 2021;124(4):686–96. https://doi.org/10.1038/s41416-020-01122-x.
Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Benson AB, Venook AP, Al-Hawary MM, et al. Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16(7):874–901. https://doi.org/10.6004/jnccn.2018.0061(ineng).
Hashiguchi Y, Muro K, Saito Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25(1):1–42. https://doi.org/10.1007/s10147-019-01485-z(ineng).
Benson AB, Venook AP, Al-Hawary MM, et al. NCCN guidelines insights: colon cancer, version 2.2018. J Natl Compr Canc Netw. 2018;16(4):359–69. https://doi.org/10.6004/jnccn.2018.0021(ineng).
Takamatsu M, Yamamoto N, Kawachi H, et al. Prediction of lymph node metastasis in early colorectal cancer based on histologic images by artificial intelligence. Sci Rep. 2022;12(1):2963. https://doi.org/10.1038/s41598-022-07038-1(ineng).
Brockmoeller S, Echle A, Ghaffari Laleh N, et al. Deep learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J Pathol. 2022;256(3):269–81. https://doi.org/10.1002/path.5831(ineng).
Kather JN, Krisam J, Charoentong P, et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16(1): e1002730. https://doi.org/10.1371/journal.pmed.1002730.
Fuhr L, Abreu M, Carbone A, et al. The Interplay between colon cancer cells and tumour-associated stromal cells impacts the biological clock and enhances malignant phenotypes. Cancers (Basel). 2019;11(7):988. https://doi.org/10.3390/cancers11070988(ineng).
Ueno H, Kanemitsu Y, Sekine S, et al. Desmoplastic pattern at the tumor front defines poor-prognosis subtypes of colorectal cancer. Am J Surg Pathol. 2017;41(11):1506–12. https://doi.org/10.1097/pas.0000000000000946(ineng).
Ozeki T, Shimura T, Ozeki T, et al. The risk analyses of lymph node metastasis and recurrence for submucosal invasive colorectal cancer: novel criteria to skip completion surgery. Cancers (Basel). 2022. https://doi.org/10.3390/cancers14030822.
Yoda Y, Ikematsu H, Matsuda T, et al. A large-scale multicenter study of long-term outcomes after endoscopic resection for submucosal invasive colorectal cancer. Endoscopy. 2013;45(9):718–24. https://doi.org/10.1055/s-0033-1344234(ineng).
Ikematsu H, Yoda Y, Matsuda T, et al. Long-term outcomes after resection for submucosal invasive colorectal cancers. Gastroenterology. 2013;144(3):551–9. https://doi.org/10.1053/j.gastro.2012.12.003.
He K, Zhang X, Ren S, et al., editors. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016
Liu L, Jiang H, He P, et al. On the variance of the adaptive learning rate and beyond. arXiv preprint arXiv:190803265. 2019
Buslaev A, Iglovikov VI, Khvedchenya E, et al. Albumentations: fast and flexible image augmentations. Information. 2020;11(2):125.
Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–6.
Acknowledgements
The authors have no conflicts of interest to disclose. This study was supported by Samsung Medical Center Grant #SMO1210071.
Funding
Samsung Medical Center,#SMO1210071,Eun Ran Kim.
Author information
Authors and Affiliations
Contributions
J.H.S.: Conception and design of the study, analysis and interpretation of data, and preparation of the manuscript; Y.H.: Conception and design of the study, performance of experiments, and preparation of the manuscript; E.R.K.: Conception and design of the study, critical revision of the manuscript for important intellectual content, and final approval of the manuscript; S–H.K., I.S.: critical revision of the manuscript for important intellectual content, and final approval of the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, J.H., Hong, Y., Kim, E.R. et al. Utility of artificial intelligence with deep learning of hematoxylin and eosin-stained whole slide images to predict lymph node metastasis in T1 colorectal cancer using endoscopically resected specimens; prediction of lymph node metastasis in T1 colorectal cancer. J Gastroenterol 57, 654–666 (2022). https://doi.org/10.1007/s00535-022-01894-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-022-01894-4