Abstract
Background
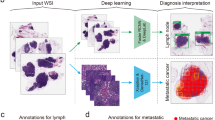
Advances in whole-slide image capture and computer image analyses using deep learning technologies have enabled the development of computer-assisted diagnostics in pathology. Herein, we built a deep learning algorithm to detect lymph node (LN) metastasis on whole-slide images of LNs retrieved from patients with gastric adenocarcinoma and evaluated its performance in clinical settings.
Methods
We randomly selected 18 patients with gastric adenocarcinoma who underwent surgery with curative intent and were positive for LN metastasis at Chiba University Hospital. A ResNet-152-based assistance system was established to detect LN metastases and to outline regions that are highly probable for metastasis in LN images. Reference standards comprising 70 LN images from two different institutions were reviewed by six pathologists with or without algorithm assistance, and their diagnostic performances were compared between the two settings.
Results
No statistically significant differences were observed between these two settings regarding sensitivity, review time, or confidence levels in classifying macrometastases, isolated tumor cells, and metastasis-negative. Meanwhile, the sensitivity for detecting micrometastases significantly improved with algorithm assistance, although the review time was significantly longer than that without assistance. Analysis of the algorithm’s sensitivity in detecting metastasis in the reference standard indicated an area under the curve of 0.869, whereas that for the detection of micrometastases was 0.785.
Conclusions
A wide variety of histological types in gastric adenocarcinoma could account for these relatively low performances; however, this level of algorithm performance could suffice to help pathologists improve diagnostic accuracy.







Similar content being viewed by others
Data availability
The datasets supporting the conclusions of this article are included within the article.
References
Kakeji Y, Ishikawa T, Suzuki S et al (2022) A retrospective 5-year survival analysis of surgically resected gastric cancer cases from the Japanese Gastric Cancer Association nationwide registry (2001–2013). Gastric Cancer. https://doi.org/10.1007/s10120-022-01317-6
Kinoshita T (2020) Minimally invasive approaches for early gastric cancer in East Asia: current status and future perspective. Transl Gastroenterol Hepatol 5:20. https://doi.org/10.21037/tgh.2019.10.08
Morton DL, Wen DR, Wong JH et al (1992) Technical details of intraoperative lymphatic mapping for early stage melanoma. Arch Surg 127(4):392–399. https://doi.org/10.1001/archsurg.1992.01420040034005
Giuliano AE, Kirgan DM, Guenther JM et al (1994) Lymphatic mapping and sentinel lymphadenectomy for breast cancer. Ann Surg 220(3):391–398. https://doi.org/10.1097/00000658-199409000-00015
Kitagawa Y, Fujii H, Mukai M et al (2000) The role of the sentinel lymph node in gastrointestinal cancer. Surg Clin North Am 80(6):1799–1809. https://doi.org/10.1016/s0039-6109(05)70262-0
Hayashi H, Ochiai T, Mori M et al (2003) Sentinel lymph node mapping for gastric cancer using a dual procedure with dye- and gamma probe-guided techniques. J Am Coll Surg 196(1):68–74. https://doi.org/10.1016/s1072-7515(02)01594-6
Natsugoe S, Arigami T, Uenosono Y et al (2017) Novel surgical approach based on the sentinel node concept in patients with early gastric cancer. Ann Gastroenterol Surg 1(3):180–185. https://doi.org/10.1002/ags3.12027
Kinami S, Fujimura T, Ojima E et al (2008) PTD classification: proposal for a new classification of gastric cancer location based on physiological lymphatic flow. Int J Clin Oncol 13(4):320–329. https://doi.org/10.1007/s10147-007-0755-x
Santos FAV, Drummond-Lage AP, Wainstein AJA et al (2020) Impact of multisection and immunohistochemistry in lymph node staging of gastric carcinoma-case series. Sci Rep 10(1):3271. https://doi.org/10.1038/s41598-020-59000-8
Zhou Y, Zhang GJ, Wang J et al (2017) Current status of lymph node micrometastasis in gastric cancer. Oncotarget 8(31):51963–51969. https://doi.org/10.18632/oncotarget.17495
Diaz Del Arco C, Ortega Medina L, Estrada Munoz L et al (2021) Pathologic lymph node staging of gastric cancer. Am J Clin Pathol 156(5):749–765. https://doi.org/10.1093/ajcp/aqab031
Srinidhi CL, Ciga O, Martel AL (2021) Deep neural network models for computational histopathology: a survey. Med Image Anal 67:101813. https://doi.org/10.1016/j.media.2020.101813
Bejnordi BE, Veta M, van Diest PJ et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22):2199–2210. https://doi.org/10.1001/jama.2017.14585
Steiner DF, MacDonald R, Liu Y et al (2018) Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol 42(12):1636–1646. https://doi.org/10.1097/PAS.0000000000001151
Japanese Gastric Cancer Association (2011) Japanese classification of gastric carcinoma. Gastric Cancer 14(2):101–112. https://doi.org/10.1007/s10120-011-0041-5
Matsushima J, Sato T, Ohnishi T et al (2022) The use of deep learning-based computer diagnostic algorithm for detection of lymph node metastases of gastric adenocarcinoma. Int J Surg Pathol. https://doi.org/10.1177/10668969221113475
He K, Zhang X, Ren S et al (2015) Deep residual learning for image recognition. Comput Sci. https://doi.org/10.48550/arXiv.1512.03385
Deng J, Dong W, Socher R et al (2009) ImageNet: a large-scale hierarchical image database. 2009 IEEE Computer Vision and Pattern Recognition (CVPR). Miami Beach, Florida. https://doi.org/10.1109/CVPR.2009.5206848
Gotoda T, Yanagisawa A, Sasako M et al (2000) Incidence of lymph node metastasis from early gastric cancer: estimation with a large number of cases at two large centers. Gastric Cancer 3(4):219–225. https://doi.org/10.1007/pl00011720
Dosovitskiy A, Beyer L, Kolesnikov A et al (2020) An image is worth 16x16words: transformers for image recognition at Scale. Comput Sci. https://doi.org/10.48550/arXiv.2010.11929
Tan M, Le QV (2021) EfficientNetV2: smaller models and faster training. Comput Sci. https://doi.org/10.48550/arXiv.2104.00298
Guan H, Liu M (2021) Domain adaptation for medical image analysis: a survey. Comput Sci. https://doi.org/10.48550/arXiv.2102.09508
Komura D, Ishikawa S (2018) Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J 16:34–42. https://doi.org/10.1016/j.csbj.2018.01.001
Liu Y, Kohlberger T, Norouzi M et al (2019) Artificial intelligence-based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med 143(7):859–868. https://doi.org/10.5858/arpa.2018-0147-OA
van der Laak JAWM, Pahlplatz MMM, Hanselaar AGJM et al (2000) Hue-saturation-density (HSD) model for stain recognition in digital images from transmitted light microscopy. Cytometry 39(4):275–284. https://doi.org/10.1002/(sici)1097-0320(20000401)39:4%3c275::Aid-cyto5%3e3.0.Co;2-8
Acknowledgements
The authors thank Kiyotaka Onodera of the Department of Pathology, Chiba University Hospital and Takuya Okamura of the Department of Pathology, Dokkyo Medical University Saitama Medical Center for their technical assistance.
Funding
This study was partially supported by the Chiba University Strategic Priority Research Promotion Program “Multimodal Medical Engineering and KAKENHI (the Grants-in-Aid for Scientific Research) (C) 20K09027.
Author information
Authors and Affiliations
Contributions
JM: Methodology, data curation, writing and original draft. TS: Methodology, software, visualization, and formal analysis. YY: supervision. HM: Project administration. SK: supervision. KM: validation. JI: Resources and writing−the review. TS: validation. AF: validation. YO: validation. TM: resources; SB: resources, supervision, and validation. HM: Resources and supervision. HH: conceptualization, formal analysis, writing, reviewing, and editing.
Corresponding author
Ethics declarations
Conflict of interest
The research leading to these results received funding from Toshiba Digital Solutions Corporation under Grant Agreement No. J20KK00041. No other relationships or activities appear to have influenced the submitted work.
Ethical approval
This study was approved by the Research Ethics Committee of the Graduate School of Medicine, Chiba University (#4122) and the Research Ethics Committee of Saitama Medical Center, Dokkyo Medical University (#21016).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Matsushima, J., Sato, T., Yoshimura, Y. et al. Clinical utility of artificial intelligence assistance in histopathologic review of lymph node metastasis for gastric adenocarcinoma. Int J Clin Oncol 28, 1033–1042 (2023). https://doi.org/10.1007/s10147-023-02356-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-023-02356-4