Abstract
Background
The mucosa-associated gut microbiota directly modulates epithelial and mucosal function. In this study, we investigated the mucosa-associated microbial community in patients with inflammatory bowel disease (IBD), using endoscopic brush samples.
Methods
A total of 174 mucus samples from 43 patients with ulcerative colitis (UC), 26 with Crohn’s disease (CD) and 14 non-IBD controls were obtained by gentle brushing of mucosal surfaces using endoscopic cytology brushes. The gut microbiome was analyzed using 16S rRNA gene sequencing.
Results
There were no significant differences in microbial structure among different anatomical sites (the ileum, cecum and sigmoid colon) within individuals. There was, however, a significant difference in microbial structure between CD, UC and non-IBD controls. The difference between CD and non-IBD controls was more marked than that between UC patients and non-IBD controls. α-Diversity was significantly lower in UC and CD patients than non-IBD controls. When comparing CD patients with non-IBD controls, the phylum Proteobacteria was significantly increased and the phyla Firmicutes and Bacteroidetes were significantly reduced. These included a significant increase in the genera Escherichia, Ruminococcus (R. gnavus), Cetobacterium, Actinobacillus and Enterococcus, and a significant decrease in the genera Faecalibacterium, Coprococcus, Prevotella and Roseburia. Comparisons between CD and UC patients revealed a greater abundance of the genera Escherichia, Ruminococcus (R. gnavus), Clostridium, Cetobacterium, Peptostreptococcus in CD patients, and the genera Faecalibacterium, Blautia, Bifidobacterium, Roseburia and Citrobacter in UC patients.
Conclusions
Mucosa-associated dysbiosis was identified in IBD patients. CD and UC may be distinguishable from the mucosa-associated microbial community structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), is a group of chronic intestinal disorders of multifactorial etiology [1,2,3]. Although the precise pathogenesis of IBD remains poorly understood, dysregulated host–microbial interactions are considered to play a role in initiating and perpetuating IBD [1, 3]. In particular, an alteration in the diversity and composition of the gut microbiome (dysbiosis), rather than the presence of specific pathogens, likely plays a critical role [3, 4]. Recent studies using deep shotgun metagenomics sequencing suggest that the metabolic activity of the gut microbiome as well as its composition may differ between subjects with IBD and healthy controls [3, 5].
The dysbiosis of IBD is characterized by a decrease in bacterial diversity and a decrease in members of the phylum Firmicutes, and concomitant expansion of the phylum Proteobacteria [6,7,8]. A reduced abundance of certain Clostridium subsets, such as cluster IV and subcluster XIVa, contributes to the decreased abundance of the phylum Firmicutes [9, 10], and an increase in the phylum Proteobacteria is represented by the expansion of Enterobacteriaceae, including E. coli. An increase in ambient oxygen levels induced by hyperemia and increased vascular and mucosal permeability is thought to be one of the mechanisms responsible for the reduction of obligate anaerobes (Clostridium groups IV or XIVa), with expansion of aerobes and facultative anaerobes (Enterobacteriaceae) [11]. Although a few studies have reported bacterial species that are potentially protective (e.g. Faecalibacterium prausnitzii) and potentially inflammatory/aggressive for IBD (e.g. adherent/invasive Escherichia coli) [3, 4], the majority of bacterial species that characterize the dysbiosis of IBD have not been fully identified.
Mucus layers play an important role in protecting intestinal epithelial cells from the gut microbes and pathogenic microorganisms [12]. In the colon, thick mucus layers, composed of a dense inner mucus layer and a loose outer mucus layer, cover luminal surfaces of the epithelium. The majority of the gut microbiota are present in the outer mucus layer, and none are present in the dense inner mucus layer [13, 14]. Inflammatory responses lead to a reduction in the number of goblet cells, reduced thickness of the mucus layer, and altered mucus composition [15, 16]. These changes enable easy contact of microbes with epithelial cells, stimulating an inflammatory response.
The gut microbiota consists of two separate populations, the luminal microbiota and the mucosa-associated microbiota [17, 18]. The mucosa-associated microbiota is believed to directly affect epithelial and mucosal function to a greater degree than luminal bacteria, and may be more deeply involved in the pathophysiology of IBD. However, studies investigating the gut microbiota of IBD have typically used fecal samples because they are easily collected, and only a few studies have investigated the mucosa-associated microbiota [17, 19, 20]. In addition, most previous studies of mucosa-associated microbiota have used samples collected by endoscopic biopsy, which is invasive and sometimes causes unexpected bleeding. Furthermore, a large part of the biopsy samples consists of human tissues with extremely small amounts of bacterial components. This may lead to misreading of 16S rRNA sequencing. In the present study, we investigated the mucosa-associated microbial profile of IBD patients using endoscopic brush samples.
Methods
Ethics
This study was approved by the ethics committee of Shiga University of Medical Science (permission no. 28-111). All patients were managed at the Division of Gastroenterology of the Shiga University of Medical Science Hospital. All participants provided written informed consent. The study was registered at the University Hospital Medical Information Network Center (UMIN 000024743).
Patients and sample collection
Twenty-six patients with CD, 43 patients with UC and 14 non-IBD participants were enrolled. Almost all patients had mild disease activity. Nineteen of the 26 patients with CD and 21 of the 43 patients with UC were in clinical remission [Crohn’s Disease Activity Index (CDAI) [21] <150 or Mayo score ≤3 [22]]. Non-IBD participants were undergoing colonoscopy for screening. A polyethylene glycol-based bowel preparation was administered. No patients were treated with antibiotics and/or probiotics. Patient characteristics are described in Table 1.
Samples were obtained by gentle brushing of mucosal surfaces, while taking care to avoid bleeding, using cytology brushes (COOK® CCB-7-240-3-S, Bloomington, IN, USA). Two samples were obtained from the ileum and sigmoid colon of each CD patient, and two samples were obtained from the cecum and sigmoid colon of each UC patient. Three samples were obtained from the ileum, cecum and sigmoid colon of each non-IBD participant. Samples were collected from normal-appearing mucosa under endoscopy, since the mucus layer is reduced in thickness or is absent at the inflamed lesion [3, 13]. A total of 174 samples were collected and analyzed.
DNA extraction from fecal samples
DNA was extracted from samples using the QIAamp UCP Pathogen Mini Kit (QIAGEN, Germantown, MD, USA) with Pathogen Lysis Tube S (QIAGEN). Samples were beaten in the presence of zirconia beads using the FastPrep FP100A Instrument (MP Biomedicals, Irvine, CA, USA). The final concentration of the DNA sample was adjusted to 10 ng/μl.
16S rRNA sequencing
16S rRNA sequencing using the MiSeq™ system (Illumina, San Diego, CA, USA) was performed according to a previously described method [23]. Briefly, the V3–V4 hypervariable regions of 16S rRNA were PCR-amplified using the universal primers 341F and 805R [24]. The amplicon was purified, and PCR was performed a second time to attach a unique combination of dual indices (I5 and I7 index). The amplicon of the second PCR was purified, and the concentration was normalized with a SequalPrep Normalization Plate Kit (Life Technologies, Tokyo, Japan). Each of the normalized amplicons was then evenly pooled and concentrated using AMPure XP beads (Beckman Coulter, Tokyo, Japan). Eleven picomoles of the library combined with phiX Control was sequenced using a 300-bp paired-end strategy according to the manufacturer’s instructions.
16S rDNA-based taxonomic analysis and statistical analysis
Processing of sequence data, including chimera check, operational taxonomic unit (OTU) definition and taxonomy assignment, was performed using QIIME version 1.9 [25], USEARCH v8.0 and UCHIME version 4.2.40 software [26]. Singletons were removed in this study. Taxonomy assignment of the resulting OTU was carried out using RDP classifier v2.10.2 with the Greengenes database (published May 2013) [27]. Statistical differences (P < 0.05) between groups in the relative abundance of bacterial phyla and genera were evaluated using Student’s paired t-tests.
α-Diversity and β-diversity
The Observed species, Chao1 and Shannon phylogenetic diversity indices were calculated by the R package “phyloseq” and statistically analyzed using a Bonferroni test. β-Diversity was estimated using the UniFrac metric to calculate the distances between the samples and was visualized by principal coordinate analysis (PCoA) and statistically analyzed using permutational multivariate analysis of variance (PERMANOVA). The figures were generated using QIIME version 1.9 software [25].
Functional changes in the microbiome
Potential changes in the microbiome at the functional level were evaluated using PICRUSt [Phylogenetic Investigation of Communities by Reconstruction of Unobserved States] software [28] and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (release 70.0) [29]. The human-specific pathways were removed from the results to focus on true bacterial pathways. The PICRUSt software uses 16S rRNA sequence profiles to estimate metagenome content based on reference bacterial genomes and the KEGG pathway database. The result was further analyzed statistically using Welch’s t test in the STAMP software program [30]. Benjamini–Hochberg-corrected P values (<0.05) were used to determine the statistical significance between the groups.
Results
A total of 3282 OTUs were detected among the 174 samples. Initially, we compared α-diversity between anatomical sites of the gastrointestinal tract using different indices [the observed species and the Chao1 index (OTU richness estimation), and the Shannon index (OTU evenness estimation)]. There was no statistically significant difference in α-diversity among the three anatomical sites (ileum, cecum and sigmoid colon) in non-IBD controls, with the exception of a significant difference in Chao1 index between the cecum and sigmoid colon (Fig. 1a). Similarly, there were no significant differences in α-diversity among different anatomical sites in the CD and UC samples (data not shown).
The mucosa-associated microbial communities of different anatomical sites. a α-Diversity indices of non-IBD controls (n = 14 for each anatomical sites). *P < 0.05 by Bonferroni test. b–d Weighted principal coordinate analysis (PCoA) plots of β-diversity measures of the microbiota communities. The same person is expressed by the same color. There was no difference in the microbial community of the different anatomical sites within non-IBD controls, UC or CD. PERMANOVA test, P = 1.0
The overall structure of the gut microbiome among different anatomical sites was evaluated using β-diversity indices calculated for weighted UniFrac distance. In non-IBD participants, principal coordinate analysis (PCoA) revealed no microbial structural differences between different anatomical sites (the ileum, cecum and sigmoid colon; PERMANOVA P = 1.0, Fig. 1b). However, the samples from the same participant tended to locate closely together and were more similar regardless of anatomical location, indicating the existence of inter-individual similarity (Fig. 1b). In CD and UC patients, there were also no structural differences in the microbial communities among anatomical sites (Fig. 1c, d), and inter-individual similarity was evident despite disease. Thus, our results indicate that there are no significant differences in microbial structure among different anatomical sites (the ileum, cecum and sigmoid colon), but the microbial structure of different anatomical sites tends to be similar in the same individual.
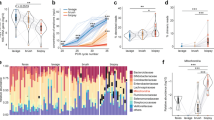
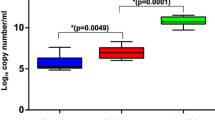
Using the unweighted and weighted UniFrac distance, we compared the overall microbial structure among non-IBD controls and UC and CD patients. As shown in Fig. 2a, the unweighted and weighted PCoA indicated significant structural differences among the three groups (PERMANOVA P = 0.001). Although overlap of all disease groups was observed to some extent, samples tended to cluster together based on disease. As shown in Fig. 2b, all indices for α-diversity were significantly lower in UC and CD patients than in non-IBD controls, although there were no significant differences between UC and CD patients. As shown in Fig. 2c, CD patients demonstrated the highest UniFrac distances (Bonferroni test for multiple comparisons), thereby indicating that the microbial differences between CD patients and non-IBD controls are more marked than those between UC patients and non-IBD controls.
Comparative analyses for the microbial community of non-IBD controls, and UC and CD patients. a Unweighted and weighted PCoA of β-diversity measures for all samples. The microbial community differed significantly between non-IBD controls and UC and CD patients (P = 0.001 by PERMANOVA test). b α-Diversity indices of non-IBD controls and UC and CD patients. *P < 0.05 by Bonferroni test. Cont. = non-IBD controls. c Weighted UniFrac distance measure. The microbial community of CD patients displayed the most dispersed clustering. *P < 0.05 by Bonferroni test
We did not collect samples directly from active lesions, since the mucus layer in such sites is reduced in thickness or is absent. Therefore, in order to evaluate the microbial communities of active lesions, we obtained brush samples from the endoscopic normal mucosa in the immediate vicinity of active lesions (UC, n = 18; CD, n = 6). As shown in Fig. 3, the weighted PCoAs and α-diversity indices revealed no significant differences in microbial communities between inactive and active lesions of UC and CD patients.
Comparative analyses for the microbial communities of inflamed and non-inflamed mucosa of UC and CD patients (upper panel). Weighted PCoA of β-diversity measures of UC and CD patients. There was no difference in the microbial community between inflamed and non-inflamed mucosa. Samples of inflamed mucosa were obtained from normal mucosa in the vicinity of active lesions. P = 1.0 by PERMANOVA test (lower panel). α-Diversity indices of the microbial communities of inflamed and non-inflamed mucosa. *P < 0.05 by Student’s t test
Differences in the gut microbial structure were taxonomically evaluated at the phylum level (Fig. 4). The abundance of the phylum Firmicutes was significantly reduced in CD patients compared to UC patients and non-IBD controls, although there was no significant difference between UC patients and non-IBD controls. The phylum Bacteroidetes was significantly reduced in CD patients. In contrast, the abundance of the phylum Proteobacteria was significantly increased in CD patients compared to UC patients and non-IBD controls. The abundance of the phylum Actinobacteria was significantly reduced in CD patients compared to UC patients.
The taxonomic changes in the microbial communities in IBD patients were evaluated at the genus level. As shown in Fig. 5a and supplementary Table 1, comparison of the microbial changes between CD patients and non-IBD controls showed a significant increase in the abundance of 15 genera and a significant decrease in 40 genera in CD patients. These included an increase in the abundance of the genera Escherichia, Ruminococcus (R. gnavus), Cetobacterium, Actinobacillus and Enterococcus, and by a decrease in the abundance of the genera Faecalibacterium, Coprococcus, Prevotella, Roseburia, Gemmiger, Alistipes, and Ruminococcus (R. bromii). The microbial changes in UC patients compared to non-IBD controls were characterized by a significant increase in eight genera and a significant decrease in 42 genera (Fig. 5b; supplementary Table 2). These included an increase in the abundance of the genera Blautia, Veillonella, Bifidobacterium, Citrobacter and Lactobacillus, and a decrease of the genera Prevotella, Coprococcus, Pseudomonas, and Alistipes. Comparison of the microbial communities between CD and UC patients revealed that the genera Escherichia, Ruminococcus (R. gnavus), Clostridium, Cetobacterium and Peptostreptococcus were significantly more abundant in CD patients, and the genera Faecalibacterium, Blautia, Bifidobacterium, Roseburia and Citrobacter were significantly more abundant in UC patients (Fig. 5c; supplementary Table 3).
Potential differences in the function of the microbiome were evaluated using PICRUSt software [28] (Fig. 6). When comparing CD patients with non-IBD controls, the proportion of genes responsible for ubiquinone and other terpenoid-quinone biosynthesis, glutathione metabolism, nitrogen metabolism, ion-coupled transporters and lipopolysaccharide biosynthesis proteins was significantly increased in CD patients. The proportion of genes responsible for bacterial chemotaxis, histidine metabolism, methane metabolism, starch and sucrose metabolism was increased in non-IBD controls. Between UC patients and non-IBD controls, the proportion of genes responsible for ion-coupled transporters, nitrogen metabolism and glutathione metabolism was increased in UC patients, while the proportion of genes responsible for bacterial translocation, transcription machinery, histidine metabolism and flagellar assembly was increased in non-IBD controls.
Discussion
The global alteration of the gut microbial community, rather than the presence of specific genera, plays an important role in the pathogenesis of IBD (dysbiosis). Previous studies have shown that the gut microbial community of IBD patients is characterized by reduced diversity and decreased abundance of the phylum Firmicutes, and a concomitant increase in the phylum Proteobacteria [6, 7, 31]. Several studies have found that the reduced diversity of the gut microbiota in IBD patients is primarily associated with the lower abundance of the phylum Firmicutes, particularly in the Clostridium cluster IV of anaerobic bacteria [4]. However, many of these studies relied on easily accessible fecal samples obtained on a single occasion, which may not properly represent mucosa-associated bacterial populations [17, 18]. The mucosa-associated microbiota is considered to affect epithelial and mucosal function to a greater degree than luminal bacteria and to contribute more significantly to the pathogenesis of IBD [13, 17, 32].
In this study, we used mucus samples obtained by gentle brushing of mucosal surfaces while avoiding bleeding under colonoscopy. Many of the previous studies of mucosa-associated gut microbiota used samples collected by mucosal biopsy [18, 20]. However, mucosal biopsy is invasive and increases the risk of unexpected bleeding. In addition, the major body of biopsy sample consists of human tissue (or cells) but contains minimal bacterial components. The advantage of the use of endoscopic brush samples is that it is non-invasive and makes it possible to avoid massive contamination of human cells. This may be advantageous in whole-genome shotgun metagenomics, since removal of human genome data is required for the metagenomic analysis of the microbiome. In order to collect sufficient mucus samples, we processed endoscopically inactive mucosa, since the mucus layer has been reported to become thinner or even disappear at the inflamed lesion [3, 13]. We also preferably selected patients in clinical remission or with mild activity, since the gut microbial structure in patients with severe clinical activity might be influenced and disturbed by various factors such as wash-away force by watery diarrhea, bleeding with serum anti-microbial proteins and increased secretion of anti-microbial peptides.
One advantage of endoscopic sampling is the ability to collect multiple samples from different anatomical sites in the same patient. From the analysis of 174 mucus samples among the different anatomical sites (ileum, cecum and sigmoid colon), we found that there was no specific microbial structure that was characteristic for each anatomical site. On the other hand, there was very little difference in the bacterial communities between the ileum, cecum and sigmoid colon in the same individual. The absence of a representative microbial structure for different anatomical sites and inter-individual uniformity were also confirmed in the samples collected from the different anatomical sites of CD and UC patients. Similar observations have been recently reported by other groups using samples collected by biopsy or other tools [17, 20]. Thus, the overall gut microbial community might be more dependent on inter-individual variation than on anatomical site.
From the analysis of a large number of mucus samples, it became clear that the gut microbial structure differed significantly among CD, UC and non-IBD controls. Comparative analysis using the weighted UniFrac distance indicated that the difference between CD and non-IBD controls was more marked than that between UC and non-IBD controls. These observations are compatible with the results of previous studies of fecal samples [33, 34]. A focal alteration of the microbial structure within the inflamed mucosa of IBD has been reported previously [35]. However, PCoA in the present study showed no clear distinction in microbial communities between active and inactive mucosa within CD (or UC) patients. There were also no significant differences in α-diversity between active and inactive mucosa. Furthermore, we failed to detect specific taxa which significantly associated with the active or inactive lesions of CD or UC patients (data not shown). These results indicate that there are no significant differences in microbial communities between inflamed and non-inflamed mucosa within CD (or UC) patients. Forbes et al. analyzed a large number of biopsy samples of IBD patients [4] and reached a similar conclusion, that localized change in the mucosa-associated microbiota does not occur in the inflamed mucosa. Thus, these findings support the notion that alteration of the gut microbial structure is not a result of mucosal inflammation, and suggest that gut dysbiosis might be an essential factor involved in the pathogenesis of IBD.
We found that dysbiosis at the phylum level was evident in CD patients but not in UC patients. When compared to non-IBD controls, the gut microbial community in CD patients was characterized by a significant decrease in the abundance of the phyla Firmicutes and Bacteroidetes, and a significant increase in the phylum Proteobacteria. This phenomenon was not observed in UC patients. These results are compatible with previous studies using fecal samples [1, 3]. However, Forbes et al. recently reported a contrasting result of taxonomic changes in the mucosa-associated microbiome of IBD using biopsy samples [20]. They demonstrated that dysbiosis at the phylum level was more evident in UC patients than CD patients. In UC patients, they observed a significant increase in the abundance of the phyla Firmicutes and Proteobacteria and a significant decrease in the phylum Bacteroidetes. These significant changes were not detected in CD patients. The reason for these contrasting observations remains unclear, but different methods for sample collection, biopsy or brushing, might be a crucial factor. A comparative study of sample collection by biopsy and brushing should be designed to determine which method yields more accurate data.
Compositional changes observed in CD patients were characterized by a significant increase in the abundance of putative aggressive bacteria, such as the genus Escherichia, the genus Ruminococcus (R. gnavus) and Fusobacterium species, combined with a significant decrease in the abundance of protective bacteria such as the genera Faecalibacterium, Coprococcus, Prevotella and Roseburia. These results are consistent with those of previous reports of fecal samples [1, 3, 4]. The pathological role of the genus Escherichia has been shown in many studies of adhesive-invasive E. coli [3, 4, 33], and R. gnavus has been reported to contribute to the pathophysiology of IBD through its mucolytic activity [35]. The genera Faecalibacterium, Coprococcus and Roseburia are the main butyrate-producing bacteria belonging to the order Clostridiales [36, 37], and play a beneficial role in maintaining gut health. Compositional changes in UC patients included an increase in the abundance of the genera Blautia, Veillonella and Bifidobacterium, and a decrease in the abundance of the genera Prevotella and Coprococcus. The genus Blautia is one of the dominant butyrate-producing bacteria in the human gut [38]. The significance of the increase in beneficial microbes (Blautia and Bifidobacterium) is unclear, although Veillonella has been reported to be potentially inflammatory [3]. When comparing the microbial composition between CD and UC patients, potentially inflammatory microbes such as the genera Escherichia and Ruminococcus (R. gnavus) were more abundant in CD patients, and potentially protective microbes such as the genera Faecalibacterium, Blautia, Bifidobacterium and Roseburia were more abundant in UC patients. This indicates that the microbial environment is more inflammatory in CD patients than in UC patients.
The functional analyses of the gut microbiome using PICRUSt software revealed that the proportion of many genes differed between IBD and non-IBD controls. Although the meaning of the change in each pathway is not clear, a significant decrease in the proportion of histidine metabolism was commonly observed in CD and UC patients. Andou et al. previously demonstrated that oral administration of histidine ameliorated intestinal inflammation in interleukin-10-deficient mice [39]. Reduced histidine metabolism in the gut microbiome in IBD patients might therefore be involved in the development of mucosal inflammation. The lipopolysaccharide (LPS) biosynthesis pathway was significantly upregulated in CD patients. LPS is a strong stimulator of the immune response derived from gram-negative bacteria [40], and this suggests an increased inflammatory status of the gut microbiome in CD patients. These are based on the indirect computational method for inferring metagenomics content, and direct metagenomic DNA sequencing will be required to confirm the findings of this study.
In conclusion, we have shown a mucosa-associated dysbiosis in IBD patients using endoscopic brush samples. Our novel findings are as follows: (a) inter-individual uniformity of the gut microbial structure at different anatomical sites of the colon and ileum, (b) no difference in the microbial structure between inflamed and non-inflamed mucosa, and (c) mucosa-associated dysbiosis more evident in CD patients than UC patients. It is possible that collection methods for mucus samples, such as biopsy or brushing, may strongly affect the results of 16S rDNA sequencing. A comparative study of sample collection methods should be considered in the future.
References
Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50:495–507.
Sun M, Wu W, Liu Z, et al. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8.
Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(327–339):e324.
Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. ILAR J. 2015;56:192–204.
Li SS, Zhu A, Benes V, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–9.
Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84.
Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–84.
Sartor RB. The intestinal microbiota in inflammatory bowel diseases. Nestle Nutr Inst Workshop Ser. 2014;79:29–39.
Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–5.
Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6.
Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(1055–1063):e1058.
McGuckin MA, Linden SK, Sutton P, et al. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–78.
Okumura R, Kurakawa T, Nakano T, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117–21.
Johansson MA, Arana-Vizcarrondo N, Biggerstaff BJ, et al. Assessing the risk of international spread of yellow fever virus: a mathematical analysis of an urban outbreak in Asuncion, 2008. Am J Trop Med Hyg. 2012;86:349–58.
Larsson JM, Karlsson H, Crespo JG, et al. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–307.
Gersemann M, Wehkamp J, Stange EF. Innate immune dysfunction in inflammatory bowel disease. J Intern Med. 2012;271:421–8.
Ringel Y, Maharshak N, Ringel-Kulka T, et al. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 2015;6:173–81.
Sartor RB. Gut microbiota: optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol. 2015;12:253–4.
Lavelle A, Lennon G, O’Sullivan O, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–61.
Forbes JD, Van Domselaar G, Bernstein CN. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm Bowel Dis. 2016;22:817–25.
Best WR, Becktel JM, Singleton JW, et al. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology. 1976;70:439–44.
Rutgeerts P, Sandborn WJ, Feagan BG, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–76.
Inoue R, Sakaue Y, Sawai S, Sawai T, Ozeki M, Romero-Pérez GA, Tsukahara T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders Bioscience. Biotechnol Biochem. 2016;80:2450–8.
Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700.
Navas-Molina JA, Peralta-Sanchez JM, Gonzalez A, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013;531:371–444.
Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72.
Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21.
Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205.
Parks DH, Tyson GW, Hugenholtz P, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4.
Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968-84.
Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–80.
Hoffmann TW, Pham HP, Bridonneau C, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J. 2016;10:460–77.
Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–22.
Png CW, Linden SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–8.
Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–61.
Duncan SH, Aminov RI, Scott KP, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–41.
Liu C, Finegold SM, Song Y, et al. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:1896–902.
Andou A, Hisamatsu T, Okamoto S, et al. Dietary histidine ameliorates murine colitis by inhibition of proinflammatory cytokine production from macrophages. Gastroenterology. 2009;136(564–574):e562.
Murdock JL, Nunez G. TLR4: the winding road to the discovery of the LPS receptor. J Immunol. 2016;197:2561–2.
Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15K08967), a Grant for the Intractable Diseases from the Ministry of Health, Labor and Welfare of Japan, and a grant from the Smoking Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Nishino, K., Nishida, A., Inoue, R. et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol 53, 95–106 (2018). https://doi.org/10.1007/s00535-017-1384-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-017-1384-4