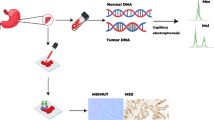
Abstract
Early detection of microsatellite instability (MSI) and microsatellite stability (MSS) is crucial in the fight against gastrointestinal (GI) cancer. MSI is a sign of genetic instability often associated with DNA repair mechanism deficiencies, which can cause (GI) cancers. On the other hand, MSS signifies genomic stability in microsatellite regions. Differentiating between these two states is pivotal in clinical decision-making as it provides prognostic and predictive information and treatment strategies. Rapid identification of MSI and MSS enables oncologists to tailor therapies more accurately, potentially saving patients from unnecessary treatments and guiding them toward regimens with the highest likelihood of success. Detecting these microsatellite status markers at an initial stage can improve patient outcomes and quality of life in GI cancer management. Our research paper introduces a cutting-edge method for detecting early GI cancer using deep learning (DL). Our goal is to identify the optimal model for GI cancer detection that surpasses previous works. Our proposed model comprises four stages: data acquisition, image processing, feature extraction, and classification. We use histopathology images from the Cancer Genome Atlas (TCGA) and Kaggle website with some modifications for data acquisition. In the image processing stage, we apply various operations such as color transformation, resizing, normalization, and labeling to prepare the input image for enrollment in our DL models. We present five different DL models, including convolutional neural networks (CNNs), a hybrid of CNNs-simple RNN (recurrent neural network), a hybrid of CNNs with long short-term memory (LSTM) (CNNs-LSTM), a hybrid of CNNs with gated recurrent unit (GRU) (CNNs-GRU), and a hybrid of CNNs-SimpleRNN-LSTM-GRU. Our empirical results demonstrate that CNNs-SimpleRNN-LSTM-GRU outperforms other models in accuracy, specificity, recall, precision, AUC, and F1, achieving an accuracy of 99.90%. Our proposed methodology offers significant improvements in GI cancer detection compared to recent techniques, highlighting the potential of DL-based approaches for histopathology data. We expect our findings to inspire future research in DL-based GI cancer detection.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer has become one of the main causes of premature death in most countries. One of the most prevalent types of cancer is gastrointestinal (GI) cancer; esophageal, gastric, colonic, and rectal tumors are all examples of GI cancers. These tumors have a variety of clinical symptoms but some common traits despite having unique but related origins [1]. GI cancers account for about 20% of all cancer diagnoses and 22.5% of cancer deaths worldwide. Gastric cancer (GC) is the third most common cause of cancer death and the fifth most common type of cancer, but esophageal cancer has a very low prevalence. After lung and breast cancer, colorectal cancer (CRC) is the third most prevalent cancer in the world, but it also ranks second in terms of cancer-related fatalities [2].
GI cancer, a collective term for a range of malignancies affecting the digestive tract, including the esophagus, stomach, liver, pancreas, colon, and rectum, remains a significant global health challenge. Among these, CRC stands as a prevalent and often lethal manifestation. The intricacies and heterogeneity of GI cancers necessitate innovative approaches for early diagnosis and tailored treatment. In recent years, the role of microsatellite instability (MSI) and microsatellite stability (MSS) [3] in these malignancies has come into focus, and the integration of artificial intelligence (AI), particularly deep learning (DL), has emerged as a promising avenue for enhancing early detection and diagnostic precision.
MSI and MSS [4,5,6] represent distinct genetic phenotypes with profound clinical implications in managing GI cancer. Microsatellites are short, repetitive DNA sequences dispersed throughout the genome, and MSI arises when these sequences exhibit instability due to defects in DNA mismatch repair (MMR) mechanisms. This instability increases the mutation burden, leading to tumors characterized by unique genetic profiles. Conversely, MSS tumors exhibit genetic stability within their microsatellite regions but may harbor distinct genetic alterations.
The differentiation between MSI and MSS in GI cancer patients [7, 8] is paramount, as it significantly informs clinical decision-making. MSI tumors are often associated with a more favorable prognosis and heightened sensitivity to immunotherapies, while MSS tumors may necessitate distinct therapeutic strategies. The accurate and timely identification of MSI or MSS status is crucial for optimizing treatment plans and ultimately improving patient outcomes.
Despite the abundance of prognostic and predictive biomarkers, high mortality rates for GI cancer patients indicate that there is still room for improvement in prognosis, opening the door to more individualized treatment approaches that could improve prognosis and/or reduce side effects. Oncology defines biomarkers as markers that also reveal the existence or absence of cancer or tumor behavior, such as therapy response or likelihood of disease recurrence. At least for the more prevalent cancer types, developing several molecular biomarkers in oncology has made it possible to treat tumors more precisely [9, 10].
Early diagnosis of GI diseases is crucial to prevent their progression into malignant diseases. There are vital indicators that aid in diagnosing GI disorders, which, if identified early, can lead to prompt treatment and a reduction in mortality rates. However, despite the availability of these diagnostic tools, the death rate due to GI diseases remains high, signifying a failure in early diagnosis and management. It is imperative to improve our diagnostic approach to ensure timely and accurate identification of GI diseases to provide appropriate care and reduce mortality rates [11]. Cutting-edge medical technologies are now employed to identify cancer at its nascent stage, with radical resection being the go-to treatment option for nearly half of all cases. Thanks to the ever-increasing number and importance of molecular biomarkers in standard clinical practice, cancer treatments are now more accurately customized to the genetic profile of a specific tumor. However, the workflow’s expenses, processing time, and tissue prerequisites are also on the rise [12, 13].
Preliminary identification of diseases like polyps and tumors through endoscopy is crucial for timely and effective treatment. However, manual diagnosis can be time-consuming and challenging, requiring extensive experience and clinical knowledge to track all video frames [14]. To overcome these difficulties, computer-aided diagnostic systems based on DL and hybrid techniques can be developed to assist doctors in making appropriate diagnoses during the early stages of a disease. By leveraging AI in the medical field, it may be possible to improve medical performance, reduce costs, and enhance the satisfaction of both patients and medical staff [15].
Biomarkers often necessitate tumor tissue in addition to standard diagnostic materials. Yet, the wealth of valuable clinical information within available tumor tissue remains largely untapped. Fortunately, modern advances in DL, an AI technology, have unlocked the ability to extract previously elusive information directly from typical cancer histology images, potentially providing valuable clinical insights [10]. With the help of artificial neural networks (ANNs), DL techniques can identify recurring patterns within complex datasets. Given the high information density of image data, it is an ideal candidate for analysis with DL techniques [16].
The growing use of AI, particularly DL-based AI, for tumor pathology is due to the development of digital pathology and advanced computer vision algorithms [17]. These DL-based algorithms can perform various tasks in tumor pathology, such as tumor diagnosis, subtyping, grading, staging, prognostic prediction, and identifying pathological features, biomarkers, and genetic changes. The use of AI in pathology improves diagnostic accuracy, reduces the workload of pathologists, and allows them to focus more on high-level decision-making tasks. Furthermore, AI is beneficial for pathologists to meet the demands of precision oncology [18, 19].
Medical computer vision has shown great promise with AI-based diagnostic support systems, specifically through convolutional neural network (CNN)-based image analysis tools. CNN-based automatic diagnosis of endoscopic findings is becoming a mainstream DL technology. This technology can help endoscopists provide more accurate diagnoses by automatically detecting and classifying endoscopic lesions [20]. However, the success of DL for endoscopy relies heavily on the availability of high-quality endoscopic images and the endoscopist’s increased understanding of the technology. DL-based image analysis has the potential to benefit numerous medical specialties that use image data, including the accurate detection of tumors and segmental organs on computer tomography (CT) images, surpassing human capabilities [21].
1.1 Motivations
Gastrointestinal (GI) cancer is a leading cause of cancer-related deaths, often going undetected in its early stages due to the limitations of traditional diagnostic methods. These methods are invasive and subjective and can lead to inconsistent diagnoses and delays in treatment. There is a pressing need for advanced diagnostic tools that enhance accuracy and speed to address these challenges.
Deep learning (DL) models present a transformative solution. By automating the analysis of complex histopathological images, DL can significantly improve the accuracy and consistency of GI cancer diagnoses. This research aims to leverage DL, particularly hybrid models combining convolutional neural networks (CNNs) and recurrent neural networks (RNNs), to capture intricate spatial and temporal patterns in histopathology images. This innovative approach aims to facilitate early detection, thereby improving patient outcomes and advancing the field of medical diagnostics.
The importance of this research lies in its potential to revolutionize GI cancer diagnosis. Early detection is crucial for effective treatment and improved survival rates. By developing a system that can accurately and quickly identify GI cancer early, we can significantly reduce the burden on healthcare systems, lower treatment costs, and, most importantly, save lives. This study not only contributes to the body of knowledge in medical AI but also has the potential for real-world impact, offering a reliable tool for clinicians in the fight against cancer.
1.2 Objectives
The primary objective of this research is to revolutionize early gastrointestinal (GI) cancer detection using advanced deep learning (DL) models. This proposed approach not only aims to provide accurate diagnoses but also to enhance patient outcomes through timely intervention. The specific goals are as follows:
-
Comprehensive preprocessing pipeline: Establish a robust preprocessing pipeline for histopathology images, integrating essential steps such as resizing, labeling, normalization, and color transformation. This ensures standardized and high-quality data preparation for subsequent analysis.
-
Efficient data handling techniques: Implement advanced data processing strategies to effectively manage and optimize large-scale histopathology image datasets. These techniques enhance computational efficiency and facilitate swift and accurate analysis.
-
Hybrid DL model design and evaluation: Develop and rigorously evaluate hybrid deep learning models that combine convolutional neural networks (CNNs) with various recurrent neural networks (RNNs) like SimpleRNN, GRU, and LSTM. These models excel in capturing nuanced spatial and temporal patterns, enhancing feature extraction and classification capabilities.
-
Robust performance metrics: Employ a comprehensive suite of performance metrics including accuracy, precision (PR), recall (RC), F1 score (F1), specificity (SP), area under the curve (AUC), and processing time. This thorough evaluation validates the effectiveness and reliability of the DL models.
-
Clinical application integration: Identify and recommend the most effective DL model for seamless integration into clinical workflows, ensuring practical applicability and enhancing diagnostic decision-making in real-world settings.
-
Proposed system overview: Provide a detailed breakdown of the proposed system, encompassing preprocessing, feature extraction, classification, and evaluation stages. Describe the architecture of each model component, elucidating their roles within the overall framework.
1.3 Contributions
This paper introduces several contributions, which are listed as follows:
-
Innovative system design: Proposes a comprehensive system for early GI cancer detection, integrating specific DL models tailored to histopathology image analysis, effectively managing a large dataset of 192,312 images.
-
Advanced preprocessing pipeline: Develops a sophisticated pipeline including data resizing, labeling, normalization, and color transformation, optimized for histopathology images. It ensures scalability and efficiency while maintaining high-quality data input for the DL models.
-
Handling large datasets: Utilizes CNNs integrated with various RNN architectures (SimpleRNN, GRU, LSTM) to address the challenge of large dataset size, enhancing processing speed and accuracy.
-
Experimental model integration: Introduces a novel integration of CNNs with RNNs for GI cancer detection. This experiment-driven approach, including custom hybrid models like CNN-SimpleRNN-LSTM-GRU, combines strengths of different networks for better accuracy and efficiency.
-
Interpretable AI: Incorporates methods such as attention mechanisms or saliency mapping to enhance model interpretability, facilitating clinical understanding and trust in the diagnostic decisions made by the models.
-
Performance optimization: Highlights hybrid models’ effectiveness in automated feature extraction and classification, focusing on improved accuracy, validation loss, and real-time processing capabilities.
-
Clinical relevance: Demonstrates practical application of hybrid DL models, supporting clinicians with accurate, timely diagnostic decisions, adaptable to different clinical settings.
-
Comprehensive evaluation and metrics: Provides a thorough evaluation using metrics like accuracy, precision(PR), recall(RC), F1 score (F1), specificity (SP), AUC, loss, and processing time, validating the approach’s effectiveness.
-
Resource management and computational efficiency: Employs strategies for optimizing computational resources and ensuring scalability, facilitating application to larger datasets or other medical image data types.
-
Future research directions: Suggests avenues for exploring molecular signatures of MSI and MSS, improving model interpretability, and testing transferability across diverse patient cohorts and datasets.
1.4 Structure
The following structure is adopted in this paper: We begin by reviewing prior research in the field in Sect. 2. Section 3 outlines different techniques for analyzing and diagnosing GI cancer. The results of experimental evaluations of the proposed systems on the dataset are presented in Sect. 4. Sections 4.4.6 and 4.5 summarize the discussion and comparison of the performance of the proposed approaches. Finally, Sect. 5 concludes the paper with recommendations for future research.
2 Literature review
This section will review several prior investigations into identifying GI diseases, especially GI cancer. We’ll also go over the techniques used to make this diagnosis and how AI and DL may aid in making the earliest and most accurate diagnoses of these disorders and assist doctors in making accurate diagnostic decisions as the following subsection:
2.1 Deep learning and neural network approaches in GI cancer diagnosis
Kather et al. [22] identified ResNet18 as the optimal neural network architecture for tumor detection in the Cancer Genome Atlas (TCGA) dataset. Their study revealed that ResNet18 exhibited several advantages, including shorter training times, superior classification performance, and a reduced risk of overfitting due to its fewer parameters. Achieving an impressive AUC score of 0.84, this study underscores the importance of selecting appropriate neural network architectures to enhance diagnostic accuracy and efficiency.
Chen et al. [23] demonstrated the efficacy of support vector machines (SVM) in automatic microsatellite instability (MSI) classification using data derived from TCGA’s stomach adenocarcinoma dataset. Their model displayed remarkable performance, with a training AUC of 0.976 and a validation AUC of 0.95. This highlights the promise of traditional machine learning (ML) techniques in MSI classification, maintaining their relevance alongside newer AI methods.
Yamashita et al. [24] introduced MSINet, a DL model for MSI detection. Evaluated on external datasets with 100 H &E-stained whole slide images (WSIs), the model achieved a net present value (NPV) of 93.7%, RC of 76.0%, SP of 66.6%, and an AUC of 0.779. Compared to expert GI pathologists, MSINet demonstrates how DL models can complement human expertise in pathology, enhancing diagnostic consistency and accuracy.
Cao et al. [25] performed a comprehensive study involving two patient cohorts from TCGA-COAD and an Asian-CRC cohort. Their innovative pathomics model, EPLA, demonstrated remarkable performance, achieving an AUC of 0.8848 in the TCGA-COAD cohort and 0.8504 in the Asian-CRC cohort. These outcomes underscore the significant potential of EPLA in clinical settings for CRC diagnosis and treatment, emphasizing the need for robust model validation across diverse populations.
Zhang et al. [26] developed a deep learning model based on preoperative CT that can predict the prognosis of patients with advanced gastric cancer. Machine learning features were extracted from the maximal tumor layer of portal vein CT images. The LASSOCox regression method was used to select features and generate labels. Then, a Cox regression model was used to integrate markers and clinicopathological information to build prediction models. Tumor segmentation was also performed to provide a more detailed analysis. It was recommended to study the possibility of verifying the results of this study retrospectively through tumor segmentation and image classification.
Padmavathi et al. [27] proposed a novel deep learning (DL) technique for classifying Gastrointestinal Disease categories from wireless capsule endoscopy (WCE) images using the mean filter to remove noise from given input images. The DenseNet-121 technique is then used to extract features such as shape and position from wireless capsule endoscopy images. The enhanced whale optimization algorithm (EWOA) is used to select the features. The experiments used the Kvasir v2 dataset and performed well regarding RC, PR, accuracy, and F1.
2.2 Incorporating transfer learning and advanced techniques
Khan et al. [28] proposed a transfer learning strategy using DL techniques and the Xception network to distinguish between MSI and MSS cancers based on histological images from formalin-fixed paraffin-embedded (FFPE) materials sourced from TCGA. Their approach yielded a notable accuracy rate of 90.17% and a test AUC of 0.932, highlighting its potential in advancing the diagnosis and treatment of these cancers. This showcases the effectiveness of transfer learning in adapting pre-trained models to new medical imaging tasks with high accuracy.
Lee et al. [29] automated MSI status classification in GC tissue slides using frozen and FFPE samples from TCGA. Their model achieved impressive AUCs of 0.893 and 0.902 for the respective sample types and an AUC of 0.874 in an external validation Asian FFPE cohort. This underscores the potential for automated MSI classification in clinical practice, highlighting the versatility and robustness of their approach.
Zhu et al. [30] developed an effective tool for predicting MSI status and assessing tumor recurrence risk in GC using preoperative Dual-Layer Computed Tomography (DLCT) scans. The study achieved excellent predictive accuracy, with an AUC of 0.879, high sensitivity, and specificity. This enhances preoperative evaluations of GC patients, demonstrating the utility of integrating advanced imaging techniques with AI for comprehensive cancer assessment.
Saldanha et al. [31] executed a retrospective study on Swarm Learning (SL) and its effectiveness in predicting molecular biomarkers in GC. Their SL-based classifier demonstrated robust performance, achieving AUCs of 0.8092 for MSI precision and 0.8372 for Epstein–Barr virus (EBV) prediction in external validation. Notably, the centralized model, trained on all datasets on a single computer, displayed comparable performance.
Qiu et al. [32] explored the relationship between MSI status and various molecular markers in CRC, developing a DL framework that predicted MSI status based solely on H &E staining images. Their framework yielded promising results, achieving an acceptable AUC of 0.809 in fivefold cross-validation for predicting MSI status using H &E images and a significantly improved AUC of 0.952 when combined with DNA methylation data. This study emphasizes the potential of DL and H &E images for MSI status prediction in CRC.
Yu et al. [33] contributed to the field by developing an innovative Deep Neural Network (DNN)-based approach to identify MSI in gastric whole slide images. Their model incorporated non-local and visual context fusion modules, resulting in an impressive accuracy rate of 96.53% and an AUC of 0.99 on the TCGA-STAD public dataset. These findings hold significant promise for advancing the diagnosis and treatment of MSI in GC.
2.3 Exploring newer AI techniques: vision transformers
Recent advancements in computer vision have seen the rise of vision transformers (ViTs) [34], which have shown significant promise in various image classification tasks [35, 36]. Here, we discuss notable applications of ViTs in medical imaging and GI cancer diagnosis:
-
Adaptive vision transformers (Adavit): Adavit offers an adaptive mechanism for Vision Transformers [37, 38], enhancing image recognition efficiency. This approach is particularly useful in medical imaging, where high accuracy and efficient computation are paramount.
-
Cross-attention multi-scale vision transformer (CrossViT): CrossViT [39, 40] improves image classification by leveraging cross-attention mechanisms and multi-scale feature extraction. This method’s ability to integrate information from different scales can be critical in identifying and classifying subtle features in medical images.
-
Evaluation of vision transformers for traffic sign classification: Although focused on traffic signs [41], this study’s principles of evaluating and optimizing Vision Transformers can be applied to medical imaging, particularly for recognizing complex patterns in histopathological slides.
-
Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling: This study [42] demonstrates ViTs’ potential in brain tumor classification, indicating their applicability to other cancers, including GI cancer, by combining multiple transformer models for improved accuracy.
Therefore, integrating vision transformers into GI cancer diagnosis could potentially enhance performance due to their ability to capture long-range dependencies and intricate patterns in medical images, making them a promising direction for future research.
2.4 Concluding remarks
From the above, we can conclude that the clinical application of digital biomarkers faces significant obstacles. However, implementing DL concepts offers considerable advantages for diagnostic and therapeutic decision-making. Despite this, there are still substantial hurdles to overcome with AI integration. These include algorithm validation and interpretability, computing systems, and skepticism from pathologists, clinicians, patients, regulators, and reimbursements. Our presentation provides an overview of how AI-based approaches can be incorporated into pathologists’ workflows. We discuss the challenges and possibilities of AI implementation in tumor pathology. Recent investigations into CNN-based image analysis in GI cancer pathology have presented encouraging findings. However, the studies were conducted in observational and retrospective contexts. Large-scale trials are necessary to evaluate performance and predict clinical usefulness. Extensive validation through large-scale trials is required before CNN-based prediction models can be authorized as medical devices.
3 The suggested approach
In this paper, a system is proposed to detect GI cancer at an early stage by utilizing specific DL models. The system comprises several key steps. Initially, the histopathology images undergo a preprocessing pipeline incorporating several techniques, including data resizing, labeling, normalization, and color transformation. The preprocessed dataset is subsequently used to develop training and testing datasets on which the recommended DL models are constructed and trained. Attention mechanisms or saliency mapping were integrated to enhance the interpretability of the models, facilitating clinical understanding and trust in their diagnostic decisions. During this study’s experiment, a significant challenge was encountered due to the large size of the data. As a result, only CNNs were employed. However, time posed another obstacle, which was overcome by combining CNNs with each type of RNN family, namely SimpleRNN, gated recurrent unit (GRU), and long short-term memory (LSTM), followed by their amalgamation. The subsequent sections detail how these DL models operate and perform automated feature extraction and classification tasks. The training accuracy and validation loss values were assessed at each period. Then, the effectiveness of the DL models was evaluated using assessment measures such as accuracy, precision(PR), recall(RC), F1 score(F1), and processing time. Figure 1 shows the general design of the proposed system for early detection of GI cancer.
3.1 The suggested neural network architecture based
3.1.1 Convolutional neural networks
Convolutional neural networks (CNNs) [43,44,45,46] represent a powerful class of DL models that have revolutionized the field of computer vision and expanded their reach into other domains. These networks are designed to process grid-like data, particularly images, with remarkable efficiency and accuracy. The fundamental concept underlying CNNs is the convolution operation, which mimics how the human visual system recognizes patterns and features. In a CNN, multiple layers of convolution, pooling, and nonlinear activation functions work together to automatically extract hierarchical features from input data, gradually learning to recognize complex patterns. This hierarchical feature extraction makes CNNs exceptionally well suited for tasks like image classification, object detection, and image segmentation. Their success has extended to diverse applications, including medical image analysis, natural language processing, and playing strategic games. CNNs continue to be a fundamental tool in AI, shaping how we process and understand visual information in the digital age. Its fundamental components and mathematical operations can be described as follows:
-
Input data: In a CNN, the input data are represented as a 3D tensor, where the dimensions are (height, width, and channels). Each dimension corresponds to the image’s height, width, and the number of color channels (e.g., red, green, blue for a color image). Mathematically, this input can be denoted as I(x, y, c), where (x, y) are the spatial coordinates, and c represents the channel.
-
Convolution operation: The core operation of a CNN is the convolution. It involves applying a set of learnable filters (kernels) to the input data to extract features. Each filter is a small grid of weights. The convolution operation at a specific location (x, y) in the feature map can be mathematically defined as follows:
$$\begin{aligned} (C * F)(x, y) = \sum \sum \sum I(x+i, y+j, c) * F(i, j, c) \end{aligned}$$(1)where (C * F)(x, y) is the result of the convolution at location (x, y), the double summation is over the spatial dimensions i and j, and the triple summation is over the input data channels c.
-
Activation function: After the convolution operation, an activation function, often ReLU (Rectified Linear Unit), is applied element-wise to introduce non-linearity. The ReLU activation function is defined as:
$$\begin{aligned} f(x) = \max (0, x) \end{aligned}$$(2)where x is the result of the convolution.
-
Pooling layer: Pooling layers are used to downsample the feature maps, reducing their spatial dimensions. Max-pooling is a common pooling operation where the maximum value within a small window is retained. Mathematically, for a pooling window size W and a feature map F, the pooling operation can be defined as:
$$\begin{aligned} (P * F)(x', y') = \max F(xW+i, yW+j) \end{aligned}$$(3)where (P * F)(x’, y’) is the result of pooling at location (x’, y’) and the summation is over the pooling window dimensions i and j.
-
Fully connected layers: The last layers of a CNN are typically fully connected layers. These layers flatten the feature maps and connect every neuron to the previous layer’s neurons. Mathematically, if N is the number of neurons in the fully connected layer, and x is a vector representing the flattened feature maps, the operation is given by:
$$\begin{aligned} \textrm{FC}(x) = W * x + b \end{aligned}$$(4)where FC(x) represents the output of the fully connected layer, W is a weight matrix, and b is a bias vector.
-
Output layer: The final layer of the CNN, known as the output layer, produces the network’s prediction or classification. Depending on the task, it might use different activation functions. For instance, image classification could employ the softmax function to assign probabilities to different classes. The softmax operation ensures that the resulting probabilities sum up to 1. Its operation is given by:
$$\begin{aligned} P(y_i) = e ^ {z{_i}} / \sum e ^ {z{_j}} \textrm{for}\, \textrm{all}\, j\, \textrm{from}\, 1\, \textrm{to}\, n \end{aligned}$$(5)where \(P(y_i)\) is the probability of the input belonging to class i, \(z_i\) is the unnormalized score (logit) associated with class i, and \(\sum\)represents the summation over all classes from 1 to n.
By stacking multiple convolutional layers, activation functions, pooling layers, and fully connected layers, a CNN, as in Fig. 2, learns to extract and represent features automatically at various abstraction levels. This mathematical framework allows CNNs to excel in various computer vision tasks, from image classification to object detection and segmentation.
3.1.2 Recurrent neural networks
Recurrent neural networks (RNNs) [47,48,49,50] are a class of ANNs designed to process sequences of data while maintaining the memory of past information. Scientifically, RNNs are characterized by their unique architecture featuring recurrent connections, which allow information to flow through loops within the network. This distinctive property makes RNNs particularly suitable for tasks involving sequential data, where the order of elements matters, such as time-series analysis, natural language processing, and speech recognition.
The fundamental component of an RNN is its hidden state, which evolves as the network processes input sequences. This hidden state acts as a form of memory, capturing and storing relevant information from previous time steps and influencing the network’s predictions at the current time step. Mathematically, RNNs are described by equations that update the hidden state and produce output predictions based on the current input and the previous hidden state.
However, traditional RNNs suffer from issues such as the vanishing gradient problem, which limits their ability to capture long-range dependencies in sequences. Advanced RNN variants, including LSTM and GRU networks, have been developed to overcome these limitations. These variants introduce gating mechanisms that regulate the flow of information within the network, allowing them to capture and retain important information over longer sequences.
The RNN family encompasses a range of architectures and variations, each with specific advantages and use cases. For instance, one-to-one RNNs handle fixed-size inputs and outputs, while one-to-many RNNs generate sequences from a single input, e.g., image captioning. Many-to-one RNNs, on the other hand, process sequences and produce a single output, e.g., sentiment analysis, and many-to-many RNNs handle sequential inputs and outputs e.g., machine translation. Understanding these variations and choosing the appropriate RNN type is crucial for tackling diverse sequence-related tasks in scientific research and practical applications. Despite their usefulness, RNNs have some limitations, such as sensitivity to hyperparameters and difficulties in parallelization. These have led to the development of alternative architectures like Transformers, which have shown exceptional performance in handling sequential data across various domains. RNNs come in various types, each tailored to specific tasks and requirements. Here are some common types of RNNs:
-
1.
SimpleRNN: A vanilla recurrent neural network (RNN), also known as a Simple RNN [44, 51, 52], is a type of ANN designed for processing data sequences. It is widely used in natural language processing, speech recognition, and time-series analysis due to its ability to capture sequential dependencies. In this explanation, we will delve into the scientific details of how a Vanilla RNN works. At its core, a Vanilla RNN, as in Fig. 3, consists of a network of interconnected neurons, with each neuron representing a hidden state at a particular time step. Let’s denote the hidden state at time step t as \(h{_t}\). The fundamental operation of a Vanilla RNN can be expressed mathematically as follows:
$$\begin{aligned} h{_t} = f(W * X{_t} + U * h{_{t-1}}) \end{aligned}$$(6)where \(X{_t}\) represents the input at time step t, W is the weight matrix connecting the input to the hidden state, U is the weight matrix connecting the previous hidden state to the current hidden state, and f is an activation function that introduces non-linearity into the network. Common choices for the activation function include the hyperbolic tangent \(\tanh\) or the \(\sigma\). The above equation shows how the current hidden state \(h{_t}\) is computed based on the input \(X{_t}\) and the previous hidden state \(h{_{t-1}}\)." This recurrent connection allows the network to maintain a memory of past information and use it to influence the current state. It is a form of weighted summation of the input and the previous hidden state, where the weights are determined by the matrices W and U. Training a Vanilla RNN involves optimizing the weights W and U to minimize a loss function that measures the difference between the predicted and target outputs. This is typically done using a technique called backpropagation through time (BPTT), which is an extension of the standard backpropagation algorithm for feedforward neural networks. BPTT computes gradients for each time step and adjusts the weights accordingly. However, Vanilla RNNs suffer from several limitations. One significant issue is the vanishing gradient problem, where gradients can become extremely small as they are propagated back during training. This can lead to difficulties in learning long-range dependencies. To address this problem, more advanced RNN variants like LSTM and Gated GRU have been developed. In conclusion, a Vanilla RNN is a fundamental type of RNN used for sequence modeling. It operates by updating a hidden state at each time step based on the input and the previous hidden state, using weighted connections. While it has been instrumental in various applications, it has limitations related to the vanishing gradient problem mitigated by more advanced RNN architectures. Understanding the mathematical operations behind Vanilla RNNs is essential for grasping the fundamentals of sequence modeling in neural networks.
-
2.
LSTM: Long short-term memory (LSTM) [53,54,55,56], a specialized RNN architecture known for its ability to capture and manage long-range dependencies in sequential data. LSTM is an advanced variant of RNNS designed to address the limitations of traditional RNNs, particularly the vanishing gradient problem, which hampers their ability to capture relationships between distant time steps in a sequence. LSTM, as in Fig. 4, achieves this by introducing a complex but effective gating mechanism composed of three key gates: the input gate (i), the forget gate (f), and the output gate (o).
-
(a)
Forget gate (f): The forget gate is crucial in deciding what information to retain or forget from the previous cell state \(C_{t-1}\). It takes as input the previous hidden state \(h_{t-1}\) and the current input \(X_t\). Through a sigmoid activation function \(\sigma\), it outputs values between 0 and 1 for each element in a vector. These values determine what parts of the previous cell state should be retained values close to 1 and what should be discarded values close to 0 as in the following equation:
$$\begin{aligned} f_t = \sigma (W_f * [h_{t-1}, X_t] + b_f) \end{aligned}$$(7)where \(W{_f}\) represents the weights associated with the forget gate and \(b{_f}\) is the bias term.
-
(b)
Input gate (i) and candidate cell state update (\(\tilde{C{_t}})\): The input gate, similar to the forget gate, utilizes the previous hidden state \(h_{t-1}\) and the current input \(X{_t}\). It employs two parts: the input gate \(i{_t}\) and the candidate cell state update \(\tilde{C{_t}}\). The input gate \(i_t\) also uses the sigmoid activation to control what new information should be added to the cell state. Simultaneously, the candidate cell state update \(\tilde{C{_t}}\) computes a new candidate cell state with a hyperbolic tangent \(\tanh\) activation function, providing a range of possible values for the new cell state as in the following equations:
$$\begin{aligned} i_t=\, & {} \sigma (W_i * [h_{t-1}, x_t] + b_i) \end{aligned}$$(8)$$\begin{aligned} \tilde{C}t=\, & {} \tanh (W_C * [h_{t-1}, x_t] + b_C) \end{aligned}$$(9) -
(c)
Cell state update (\(C_{t}\)): The cell state \(C_{t}\) is updated by combining information from the previous cell state that was determined to be relevant by the forget gate \(f_{t}\) and the new candidate values \(\tilde{C_{t}}\) determined by the input gate \(i_{t}\) as in the following equation:
$$\begin{aligned} C_t =\, f_t * C_{t-1} + i_t * \tilde{C_{t}} \end{aligned}$$(10) -
(d)
Output gate (o) and hidden state (h): The output gate (o) decides which parts of the cell state should be exposed as the hidden state \(h_{t}\) and propagated to the next time step. Like the input and forget gates, it uses the previous hidden state \(h_{t-1}\) and the current input \(X_{t}\), employing a sigmoid activation for gating and a tanh activation to compute the new hidden state as in the following equations:
$$\begin{aligned} o_t=\, & {} \sigma (W_o * [h_{t-1}, x_t] + b_o) \end{aligned}$$(11)$$\begin{aligned} h_t=\, & {} o_t * \textrm{tanh}(C_t) \end{aligned}$$(12)
The LSTM architecture incorporates these gates and states to selectively retain, update, and expose information over time. It is exceptionally suited for modeling complex sequential data with long-term dependencies. LSTMs have proved highly effective in various applications, including natural language processing, speech recognition, and time-series forecasting, where understanding intricate temporal relationships is paramount. The gating mechanism of LSTMs addresses the vanishing gradient problem, enabling the network to learn and remember important information over extended sequences, making them a powerful tool in deep learning.
-
3.
GRU: Gated recurrent unit (GRU) [57,58,59], another popular RNN architecture that shares similarities with LSTM but has a simpler structure while retaining the ability to capture long-range dependencies in sequential data. GRU, like LSTM, is designed to mitigate the vanishing gradient problem and improve the modeling of temporal relationships. It is also effective at preventing exploding gradients to some extent. Exploding gradients occur when the gradients become exceedingly large during training, which can lead to numerical instability and slow convergence. GRUs, as in Fig. 5, through their gating mechanism, help stabilize the gradients and prevent them from becoming excessively large. It achieves this through two gates: the update gate (z) and the reset gate (r).
-
(a)
Update gate (z) and reset gate (r): At the heart of the GRU architecture are the update and reset gates. These gates are responsible for controlling the flow of information within the unit. The update gate determines what portion of the previous hidden state \(h_{t-1}\) should be preserved. In contrast, the reset gate decides what information from the previous hidden state and current input \(X_{t}\) should be forgotten. Both gates are computed using sigmoid activation functions as in the following equations:
$$\begin{aligned} z_t = \sigma (W_z * [h_{t-1}, x_t]) \end{aligned}$$(13)$$\begin{aligned} r_t = \sigma (W_r * [h_{t-1}, x_t]) \end{aligned}$$(14)where \(W_{z}\) and \(W_{r}\)represent the weight matrices associated with the update and reset gates.
-
(b)
Candidate hidden state \(\tilde{h_{t}}\): The candidate hidden state \(\tilde{h_{t}}\)is a temporary representation of the information that might be included in the current hidden state. It is computed by applying the reset gate \(r_{t}\) to the previous hidden state \(h{t-1}\) and combining it with the current input \(x_{t}\) through a hyperbolic tangent tanh activation as in the following equation:
$$\begin{aligned} \tilde{h_t} = \textrm{tan}h(W_h * [r_t * h{t-1}, x_t]) \end{aligned}$$(15)where \(W_h\) represents the weight matrix associated with the candidate’s hidden state.
-
(c)
Update of the hidden state \(h_t\): The final step in the GRU’s operations involves updating the hidden state \(h_t\) by taking into account the update gate \(z_t\) and the candidate hidden state \(\tilde{h_t}\) as in the following equation:
$$\begin{aligned} h_t = (1 - z_t) * h_{t-1} + z_t * \tilde{h_t} \end{aligned}$$(16)This equation combines the previous hidden state \(h_{t-1}\) with the candidate hidden state \(\tilde{h_t}\) based on the values of the update gate \(z_t\). If the update gate is close to 1, it means that most of the previous hidden state will be preserved, and if it is close to 0, it means that most of the new information will be retained.
In summary, the GRU is an RNN architecture that utilizes update and reset gates to regulate the flow of information within the network. While simpler than LSTM’s, its design still enables it to capture long-range dependencies in sequential data effectively. GRUs have gained popularity due to their computational efficiency and competitive performance in various applications, including natural language processing, speech recognition, and time-series forecasting. The mathematical operations described here are the key components of the GRU’s functionality, allowing it to model complex temporal relationships in sequential data.
3.2 The suggested DL models
3.2.1 The suggested CNN model
In this paper, the suggested CNN model in Fig. 6 involves 11 layers. It consists of several convolutional and pooling layers to discover the preprocessed images’ features and perform the classification task. The model is created sequentially, meaning you can add layers sequentially, one after the other. The following layers make up the proposed CNNs model:
-
Convolutional layers:
-
First convolutional layer: Let the input image be \(\textbf{I} \in \mathbb {R}^{65 \times 65 \times 3}\). The first convolutional layer applies 16 filters \(\textbf{W}_1 \in \mathbb {R}^{3 \times 3 \times 3 \times 16}\) with ReLU activation. The output \(\textbf{O}_1\) is given by:
$$\begin{aligned} \textbf{O}_1 = \text {ReLU}(\textbf{I} * \textbf{W}_1 + \textbf{b}_1) \end{aligned}$$where \(*\) denotes the convolution operation, and \(\textbf{b}_1 \in \mathbb {R}^{16}\) is the bias term.
-
Subsequent convolutional layers: For the \(i\)-th convolutional layer (\(i=2,3,4\)), each with \(f_i\) filters of size \(3 \times 3\), the output \(\textbf{O}_i\) is:
$$\begin{aligned} \textbf{O}_i = \text {ReLU}(\textbf{O}_{i-1} * \textbf{W}_i + \textbf{b}_i) \end{aligned}$$where \(\textbf{W}_i \in \mathbb {R}^{3 \times 3 \times f_{i-1} \times f_i}\) and \(\textbf{b}_i \in \mathbb {R}^{f_i}\). The number of filters \(f_i\) increases as follows: \(f_2 = 32\), \(f_3 = 64\), and \(f_4 = 128\).
-
-
Max-pooling layers: Each max-pooling layer reduces the spatial dimensions of the feature maps. If \(\textbf{P}_i\) denotes the output of the max-pooling operation after the \(i\)-th convolutional layer, it is given by:
$$\begin{aligned} \textbf{P}_i = \text {MaxPool}(\textbf{O}_i, (2, 2)) \end{aligned}$$where \(\text {MaxPool}\) represents the max-pooling operation with a pool size of \(2 \times 2\).
-
Flatten layer: The output of the final max-pooling layer \(\textbf{P}_4\) is flattened to form a 1D vector \(\textbf{F}\):
$$\begin{aligned} \textbf{F} = \text {Flatten}(\textbf{P}_4) \end{aligned}$$ -
Fully connected (dense) layers:
-
First dense layer: The first dense layer with 512 units applies the ReLU activation function. The output \(\textbf{D}_1\) is:
$$\begin{aligned} \textbf{D}_1 = \text {ReLU}(\textbf{F} \cdot \textbf{W}_5 + \textbf{b}_5) \end{aligned}$$where \(\textbf{W}_5 \in \mathbb {R}^{n \times 512}\) (with \(n\) being the length of the flattened vector) and \(\textbf{b}_5 \in \mathbb {R}^{512}\).
-
Second dense layer: The second dense layer, with 2 units and softmax activation, produces the final class probabilities \(\textbf{D}_2\):
$$\begin{aligned} \textbf{D}_2 = \text {softmax}(\textbf{D}_1 \cdot \textbf{W}_6 + \textbf{b}_6) \end{aligned}$$where \(\textbf{W}_6 \in \mathbb {R}^{512 \times 2}\) and \(\textbf{b}_6 \in \mathbb {R}^{2}\).
-
The architecture described ensures the effective extraction of spatial features through convolutional layers, dimensionality reduction via max-pooling layers, and final classification using fully connected layers.
The reason for selecting CNNs stems from their well-established efficacy in handling image processing tasks. Their ability to capture spatial hierarchies in images through convolutional and pooling layers provides an effective means to discern complex textures and patterns. When considering histopathology data, these images often contain intricate spatial structures and features that require sophisticated feature extraction. CNNs excel in extracting these relevant spatial features, making them highly suitable for initial processing and feature extraction in GI cancer classification.
3.2.2 The hybrid CNN-SimpleRNN model
In this paper, a hybrid model that combines CNNs with SimpleRNN, as in Fig. 7, can be a valuable tool in image classification, even when the data are not inherently sequential. While CNNs are typically used for processing spatial information in images, and SimpleRNN is known for handling sequential data, this combination introduces a unique capability to capture non-sequential patterns and relationships. When applied to image classification with non-sequential data, this hybrid model can have a profound impact on both accuracy and training time.
The description of this hybrid model is presented in Fig. 8 and in the following points:
-
Input layer: The input image \(\textbf{I} \in \mathbb {R}^{50 \times 50 \times 3}\) represents a 50x50 pixel image with three color channels (RGB).
-
CNN layers:
-
First CNN layer:
$$\begin{aligned} \textbf{O}_1 = \text {ReLU}(\textbf{I} * \textbf{W}_1 + \textbf{b}_1) \end{aligned}$$where \(\textbf{W}_1 \in \mathbb {R}^{3 \times 3 \times 3 \times 64}\) are the filters and \(\textbf{b}_1 \in \mathbb {R}^{64}\) is the bias.
-
First max-pooling layer:
$$\begin{aligned} \textbf{P}_1 = \text {MaxPool}(\textbf{O}_1, (2, 2)) \end{aligned}$$ -
Second CNN layer:
$$\begin{aligned} \textbf{O}_2 = \text {ReLU}(\textbf{P}_1 * \textbf{W}_2 + \textbf{b}_2) \end{aligned}$$where \(\textbf{W}_2 \in \mathbb {R}^{3 \times 3 \times 64 \times 128}\) and \(\textbf{b}_2 \in \mathbb {R}^{128}\).
-
Second max-pooling layer:
$$\begin{aligned} \textbf{P}_2 = \text {MaxPool}(\textbf{O}_2, (2, 2)) \end{aligned}$$ -
Third CNN layer:
$$\begin{aligned} \textbf{O}_3 = \text {ReLU}(\textbf{P}_2 * \textbf{W}_3 + \textbf{b}_3) \end{aligned}$$where \(\textbf{W}_3 \in \mathbb {R}^{3 \times 3 \times 128 \times 512}\) and \(\textbf{b}_3 \in \mathbb {R}^{512}\).
-
Third max-pooling layer:
$$\begin{aligned} \textbf{P}_3 = \text {MaxPool}(\textbf{O}_3, (2, 2)) \end{aligned}$$
-
-
Flatten layer: The output of the final max-pooling layer is flattened into a 1D vector \(\textbf{F}\):
$$\begin{aligned} \textbf{F} = \text {Flatten}(\textbf{P}_3) \end{aligned}$$ -
SimpleRNN integration:
-
The flattened vector \(\textbf{F}\) is reshaped for RNN processing. Let \(\textbf{F}_{\text {exp}}\) be the expanded vector along the time dimension.
$$\begin{aligned} \textbf{F}_{\text {exp}} = \text {Reshape}(\textbf{F}, \text {new\_shape}) \end{aligned}$$ -
The SimpleRNN layer processes the expanded vector:
$$\begin{aligned} \textbf{R} = \text {SimpleRNN}(\textbf{F}_{\text {exp}}, \textbf{W}_R, \textbf{b}_R) \end{aligned}$$where \(\textbf{W}_R\) and \(\textbf{b}_R\) are the weights and biases of the SimpleRNN layer, respectively.
-
-
Combination layer: The outputs of the CNN and SimpleRNN layers are concatenated:
$$\begin{aligned} \textbf{C} = \text {Concatenate}([\textbf{F}, \textbf{R}]) \end{aligned}$$ -
Output layer: The final dense layer with 2 units and softmax activation for classification:
$$\begin{aligned} \textbf{Y} = \text {softmax}(\textbf{C} \cdot \textbf{W}_O + \textbf{b}_O) \end{aligned}$$where \(\textbf{W}_O \in \mathbb {R}^{d \times 2}\) and \(\textbf{b}_O \in \mathbb {R}^{2}\).
Finally, the use of this hybrid architecture not only improves the model’s classification capabilities in the presence of non-sequential data patterns but can also have an impact on reducing the time required for both training and testing, as it is explained in Sect. 4.4.
The rationale for choosing SimpleRNNs in this hybrid model lies in their ability to capture temporal dependencies in data. This integration provides temporal context when combined with CNNs, even in non-sequential histopathology data. In alignment with histopathology data, the hybrid model’s approach delivers a nuanced understanding of both spatial and temporal patterns. This enables a more precise and adaptable classification of gastrointestinal (GI) cancer, contributing to a more comprehensive interpretation of complex histopathological images.
3.2.3 The hybrid CNN-LSTM model
A hybrid model that combines CNNs with LSTM networks, as in Fig. 9 provides a robust solution for image classification, particularly when dealing with non-sequential data. While CNNs are traditionally employed for image feature extraction, LSTMs are well suited for capturing sequential dependencies and patterns, such as those in image data. This hybrid approach enhances the model’s ability to recognize non-sequential features and relationships within the data while also introducing the potential to reduce the time required for both training and testing.
The description of this hybrid model is presented in Fig. 10 and as in the following points:
-
Time-distributed CNN layers:
-
First time-distributed CNN layer:
$$\begin{aligned} \textbf{O}_1^{(t)} = \text {ReLU}(\textbf{I}^{(t)} * \textbf{W}_1 + \textbf{b}_1) \end{aligned}$$where \(\textbf{W}_1 \in \mathbb {R}^{3 \times 3 \times 3 \times 64}\) are the filters, \(\textbf{b}_1 \in \mathbb {R}^{64}\) is the bias, and \(t\) represents the time step.
-
First time-distributed max-pooling layer:
$$\begin{aligned} \textbf{P}_1^{(t)} = \text {MaxPool}(\textbf{O}_1^{(t)}, (2, 2)) \end{aligned}$$ -
Second time-distributed CNN layer:
$$\begin{aligned} \textbf{O}_2^{(t)} = \text {ReLU}(\textbf{P}_1^{(t)} * \textbf{W}_2 + \textbf{b}_2) \end{aligned}$$where \(\textbf{W}_2 \in \mathbb {R}^{3 \times 3 \times 64 \times 128}\) and \(\textbf{b}_2 \in \mathbb {R}^{128}\).
-
Second time-distributed max-pooling layer:
$$\begin{aligned} \textbf{P}_2^{(t)} = \text {MaxPool}(\textbf{O}_2^{(t)}, (2, 2)) \end{aligned}$$ -
Third time-distributed CNN layer:
$$\begin{aligned} \textbf{O}_3^{(t)} = \text {ReLU}(\textbf{P}_2^{(t)} * \textbf{W}_3 + \textbf{b}_3) \end{aligned}$$where \(\textbf{W}_3 \in \mathbb {R}^{3 \times 3 \times 128 \times 128}\) and \(\textbf{b}_3 \in \mathbb {R}^{128}\).
-
Third time-distributed max-pooling Layer:
$$\begin{aligned} \textbf{P}_3^{(t)} = \text {MaxPool}(\textbf{O}_3^{(t)}, (2, 2)) \end{aligned}$$
-
-
Time-distributed flatten layer: The output of the final max-pooling layer is flattened into a 1D vector for each time step \(t\):
$$\begin{aligned} \textbf{F}^{(t)} = \text {Flatten}(\textbf{P}_3^{(t)}) \end{aligned}$$ -
LSTM layer: The sequence of flattened vectors \(\{\textbf{F}^{(t)}\}\) is processed by the LSTM layer:
$$\begin{aligned} \textbf{H}_t = \text {LSTM}(\textbf{F}^{(t)}, \textbf{H}_{t-1}, \textbf{C}_{t-1}, \textbf{W}_L, \textbf{b}_L) \end{aligned}$$where \(\textbf{H}_t\) and \(\textbf{C}_t\) are the hidden and cell states at time step \(t\), and \(\textbf{W}_L\) and \(\textbf{b}_L\) are the weights and biases of the LSTM layer.
-
Output layer: The final dense layer with 2 units and softmax activation for classification:
$$\begin{aligned} \textbf{Y} = \text {softmax}(\textbf{H}_T \cdot \textbf{W}_O + \textbf{b}_O) \end{aligned}$$where \(\textbf{W}_O \in \mathbb {R}^{256 \times 2}\) and \(\textbf{b}_O \in \mathbb {R}^{2}\).
Integrating an LSTM network into the traditional CNN architecture offers the model a better understanding of non-sequential data patterns and relationships. While the precise reduction in training and testing time can vary based on factors like dataset size, complexity, and hardware resources, this hybrid approach can potentially enhance the efficiency and accuracy of the image classification process. Combining the strengths of CNNs for spatial feature extraction and LSTMs for sequential dependencies, this model provides a robust solution for image classification tasks with non-sequential data.
The choice of LSTMs in this hybrid model is based on their advanced capability to capture long-term dependencies and address the vanishing gradient problem. LSTMs are well suited for complex temporal feature extraction. When combined with CNNs, LSTMs can detect intricate spatial and temporal relationships in histopathology images, contributing to the precise classification of GI cancer.
3.2.4 The hybrid CNN-GRU model
A hybrid model that combines CNNs with GRU, as in Fig. 11, presents a powerful approach to image classification, particularly when dealing with non-sequential data. CNNs excel at extracting spatial features from images, while GRUs, a type of RNN, can capture temporal dependencies in data. This hybrid model not only enhances the recognition of non-sequential patterns and relationships within the data but also can potentially reduce the time required for training and testing.
The description of this hybrid model is presented in Fig. 12 and in the following points:
-
Input layer: The model processes input images of dimensions (50, 50, 3), representing a \(50 \times 50\) pixel image with three color channels (RGB).
-
CNN layers:
-
First CNN layer:
$$\begin{aligned} \textbf{O}_1 = \text {ReLU}(\textbf{I} * \textbf{W}_1 + \textbf{b}_1) \end{aligned}$$where \(\textbf{W}_1 \in \mathbb {R}^{3 \times 3 \times 3 \times 16}\) are the filters, and \(\textbf{b}_1 \in \mathbb {R}^{16}\) is the bias.
-
First max-pooling layer:
$$\begin{aligned} \textbf{P}_1 = \text {MaxPool}(\textbf{O}_1, (2, 2)) \end{aligned}$$ -
Second CNN layer:
$$\begin{aligned} \textbf{O}_2 = \text {ReLU}(\textbf{P}_1 * \textbf{W}_2 + \textbf{b}_2) \end{aligned}$$where \(\textbf{W}_2 \in \mathbb {R}^{3 \times 3 \times 16 \times 32}\) and \(\textbf{b}_2 \in \mathbb {R}^{32}\).
-
Second max-pooling layer:
$$\begin{aligned} \textbf{P}_2 = \text {MaxPool}(\textbf{O}_2, (2, 2)) \end{aligned}$$ -
Third CNN layer:
$$\begin{aligned} \textbf{O}_3 = \text {ReLU}(\textbf{P}_2 * \textbf{W}_3 + \textbf{b}_3) \end{aligned}$$where \(\textbf{W}_3 \in \mathbb {R}^{3 \times 3 \times 32 \times 64}\) and \(\textbf{b}_3 \in \mathbb {R}^{64}\).
-
Third max-pooling layer:
$$\begin{aligned} \textbf{P}_3 = \text {MaxPool}(\textbf{O}_3, (2, 2)) \end{aligned}$$ -
Fourth CNN layer:
$$\begin{aligned} \textbf{O}_4 = \text {ReLU}(\textbf{P}_3 * \textbf{W}_4 + \textbf{b}_4) \end{aligned}$$where \(\textbf{W}_4 \in \mathbb {R}^{3 \times 3 \times 64 \times 128}\) and \(\textbf{b}_4 \in \mathbb {R}^{128}\).
-
Fourth max-pooling layer:
$$\begin{aligned} \textbf{P}_4 = \text {MaxPool}(\textbf{O}_4, (2, 2)) \end{aligned}$$
-
-
Flatten layer: The output of the final max-pooling layer is flattened into a 1D vector:
$$\begin{aligned} \textbf{F} = \text {Flatten}(\textbf{P}_4) \end{aligned}$$ -
GRU integration:
-
Expanding time dimension: The flattened vector \(\textbf{F}\) is expanded along the time dimension (axis=1) to prepare the data for the GRU layer:
$$\begin{aligned} \textbf{F}_{\text {expanded}} = \text {ExpandDims}(\textbf{F}, \text {axis}=1) \end{aligned}$$ -
GRU layer: A GRU layer with 64 units processes the expanded vector:
$$\begin{aligned} \textbf{H}_t = \text {GRU}(\textbf{F}_{\text {expanded}}, \textbf{H}_{t-1}, \textbf{W}_G, \textbf{b}_G) \end{aligned}$$where \(\textbf{H}_t\) is the hidden state at time step \(t\), and \(\textbf{W}_G\) and \(\textbf{b}_G\) are the weights and biases of the GRU layer.
-
-
Output layer: The final dense layer with 2 units and softmax activation for classification:
$$\begin{aligned} \textbf{Y} = \text {softmax}(\textbf{H}_T \cdot \textbf{W}_O + \textbf{b}_O) \end{aligned}$$where \(\textbf{W}_O \in \mathbb {R}^{64 \times 2}\) and \(\textbf{b}_O \in \mathbb {R}^{2}\).
Integrating a GRU network into the traditional CNN architecture gives the model a broader understanding of non-sequential data patterns and relationships. While the exact reduction in training and testing time will depend on factors like dataset size, complexity, and available hardware resources, this hybrid approach can potentially enhance the efficiency and accuracy of the image classification process. Combining the strengths of CNNs for spatial feature extraction and GRUs for temporal dependencies, this model offers a robust solution for image classification tasks with non-sequential data.
The reason for selecting GRUs in this model is their computational efficiency and ability to detect temporal dependencies within data. Compared to LSTMs, GRUs offer a more streamlined approach while still maintaining strong performance. In alignment with histopathology data, GRUs can work in conjunction with CNNs to recognize temporal patterns alongside spatial features. This makes them suitable for identifying complex relationships in histopathology images that require a nuanced understanding of temporal context.
3.2.5 The hybrid CNN-SimpleRNN-LSTM-GRU model:
A hybrid model that combines CNNs with multiple RNN layers as in Fig. 13, including SimpleRNN, LSTM, and GRU, presents a comprehensive and versatile approach to image classification, particularly when dealing with non-sequential data. While CNNs excel at extracting spatial features from images, RNNs are well suited to capture temporal and sequential dependencies within data. This hybrid model offers an extensive understanding of non-sequential data patterns and relationships, and it can significantly affect the efficiency of both training and testing processes.
The description of the hybrid model is presented in Fig. 14 and in the following points:
-
Input layer: The model processes input images with dimensions of (50, 50, 3), representing a \(50 \times 50\) pixel image with three color channels (RGB).
-
CNN layers: The model begins with four CNN layers. Each layer applies a 2D convolution operation followed by a max-pooling operation:
-
First CNN layer:
$$\begin{aligned} \textbf{O}_1 = \text {ReLU}(\textbf{I} * \textbf{W}_1 + \textbf{b}_1) \end{aligned}$$where \(\textbf{W}_1 \in \mathbb {R}^{3 \times 3 \times 3 \times 16}\) are the filters and \(\textbf{b}_1 \in \mathbb {R}^{16}\) is the bias.
-
First max-pooling layer:
$$\begin{aligned} \textbf{P}_1 = \text {MaxPool}(\textbf{O}_1, (2, 2)) \end{aligned}$$ -
Subsequent CNN and max-pooling layers follow a similar structure with increased filter numbers (32, 64, and 128).
-
Second CNN layer:
$$\begin{aligned} \textbf{O}_2 = \text {ReLU}(\textbf{P}_1 * \textbf{W}_2 + \textbf{b}_2) \end{aligned}$$where \(\textbf{W}_2 \in \mathbb {R}^{3 \times 3 \times 16 \times 32}\) and \(\textbf{b}_2 \in \mathbb {R}^{32}\).
-
Second max-pooling layer:
$$\begin{aligned} \textbf{P}_2 = \text {MaxPool}(\textbf{O}_2, (2, 2)) \end{aligned}$$ -
Third CNN layer:
$$\begin{aligned} \textbf{O}_3 = \text {ReLU}(\textbf{P}_2 * \textbf{W}_3 + \textbf{b}_3) \end{aligned}$$where \(\textbf{W}_3 \in \mathbb {R}^{3 \times 3 \times 32 \times 64}\) and \(\textbf{b}_3 \in \mathbb {R}^{64}\).
-
Third max-pooling layer:
$$\begin{aligned} \textbf{P}_3 = \text {MaxPool}(\textbf{O}_3, (2, 2)) \end{aligned}$$ -
Fourth CNN layer:
$$\begin{aligned} \textbf{O}_4 = \text {ReLU}(\textbf{P}_3 * \textbf{W}_4 + \textbf{b}_4) \end{aligned}$$where \(\textbf{W}_4 \in \mathbb {R}^{3 \times 3 \times 64 \times 128}\) and \(\textbf{b}_4 \in \mathbb {R}^{128}\).
-
Fourth max-pooling layer:
$$\begin{aligned} \textbf{P}_4 = \text {MaxPool}(\textbf{O}_4, (2, 2)) \end{aligned}$$
-
-
Flatten layer: The output of the final max-pooling layer is flattened into a 1D vector:
$$\begin{aligned} \textbf{F} = \text {Flatten}(\textbf{P}_4) \end{aligned}$$ -
SimpleRNN integration:
-
Expanding time dimension: The flattened vector \(\textbf{F}\) is expanded along the time dimension (axis=1) to prepare the data for the SimpleRNN layer:
$$\begin{aligned} \textbf{F}_{\text {expanded}} = \text {ExpandDims}(\textbf{F}, \text {axis}=1) \end{aligned}$$ -
SimpleRNN layer: A SimpleRNN layer with 64 units processes the expanded vector:
$$\begin{aligned} \textbf{H}^{\text {SimpleRNN}}_t = \text {SimpleRNN}(\textbf{F}_{\text {expanded}}, \textbf{H}_{t-1}, \textbf{W}_{SR}, \textbf{b}_{SR}) \end{aligned}$$where \(\textbf{H}^{\text {SimpleRNN}}_t\) is the hidden state at time step \(t\), and \(\textbf{W}_{SR}\) and \(\textbf{b}_{SR}\) are the weights and biases of the SimpleRNN layer.
-
-
LSTM integration:
-
A separate sequence of the expanded data is input to an LSTM layer with 64 units:
$$\begin{aligned} \textbf{H}^{\text {LSTM}}_t = \text {LSTM}(\textbf{F}_{\text {expanded}}, \textbf{H}_{t-1}, \textbf{C}_{t-1}, \textbf{W}_{LSTM}, \textbf{b}_{LSTM}) \end{aligned}$$where \(\textbf{H}^{\text {LSTM}}_t\) is the hidden state, \(\textbf{C}_{t-1}\) is the cell state at time step \(t-1\), and \(\textbf{W}_{LSTM}\) and \(\textbf{b}_{LSTM}\) are the weights and biases of the LSTM layer.
-
-
GRU integration:
-
Another sequence of the expanded data is input to a GRU layer with 64 units:
$$\begin{aligned} \textbf{H}^{\text {GRU}}_t = \text {GRU}(\textbf{F}_{\text {expanded}}, \textbf{H}_{t-1}, \textbf{W}_{GRU}, \textbf{b}_{GRU}) \end{aligned}$$where \(\textbf{H}^{\text {GRU}}_t\) is the hidden state at time step \(t\), and \(\textbf{W}_{GRU}\) and \(\textbf{b}_{GRU}\) are the weights and biases of the GRU layer.
-
-
Concatenation layer: The outputs of the SimpleRNN, LSTM, and GRU layers are concatenated:
$$\begin{aligned} \textbf{H}_{\text {concat}} = \text {Concatenate}([\textbf{H}^{\text {SimpleRNN}}_T, \textbf{H}^{\text {LSTM}}_T, \textbf{H}^{\text {GRU}}_T]) \end{aligned}$$ -
Output layer: A dense output layer with 2 units and softmax activation for classification:
$$\begin{aligned} \textbf{Y} = \text {softmax}(\textbf{H}_{\text {concat}} \cdot \textbf{W}_O + \textbf{b}_O) \end{aligned}$$where \(\textbf{W}_O \in \mathbb {R}^{192 \times 2}\) and \(\textbf{b}_O \in \mathbb {R}^{2}\).
Integrating SimpleRNN, LSTM, and GRU networks alongside the traditional CNN architecture allows the model to analyze non-sequential data patterns and relationships comprehensively. While the exact reduction in training and testing time will depend on factors like dataset size, complexity, and available hardware resources, this hybrid approach can significantly improve the efficiency and accuracy of the image classification process. Combining the strengths of CNNs for spatial feature extraction with multiple RNN architectures for temporal dependencies, this model offers a powerful solution for image classification tasks with non-sequential data.
The rationale for choosing this hybrid model stems from its capacity to blend the spatial feature extraction prowess of convolutional neural networks (CNNs) with the temporal feature extraction capabilities of SimpleRNN, LSTM, and GRU. This integration provides a robust framework that can accommodate diverse temporal and spatial features found in histopathology images, thereby enhancing the precision of the classification process. In terms of alignment with histopathology data, the hybrid model’s combined approach offers a comprehensive insight into both spatial and temporal patterns. Such a dual understanding facilitates a more accurate and adaptable classification of gastrointestinal (GI) cancer in histopathological images, offering a deeper and more nuanced interpretation of complex biological features.
4 Exploratory outcomes and analysis
This section presents the results of our proposed models and an evaluation of the datasets used for training and testing. The final results were obtained by averaging all the evaluation metrics, and we have also recorded the time taken for training and testing. To assess the performance of our models, we have included the datasets used in the Sect. 4.1. We have also provided workplace characteristics in Sect. 4.2 and performance indicators in Sect. 4.3. Finally, Sect. 4.4 presents a comparative analysis.
4.1 Dataset characterization
The proposed and state-of-the-art models are assessed in this paper using histopathology data. All slides of this data are available at.Footnote 1 It is accessible through Zenodo atFootnote 2 and also through Kaggle at.Footnote 3 The dataset, which includes 192312 histopathology images, is divided into two classes: 75039 MSIMUT images and 117273 MSS images, as demonstrated in Figs. 15 and 16.
4.1.1 Data preprocessing
The data undergo a series of preprocessing steps, which include resizing, color transformation, and normalization, followed by a labeling process. These measures ensure that the data are in a suitable format for analysis or modeling.
Image preprocessing [60, 61, 61, 62] refers to a set of techniques and operations that are applied to digital images to prepare them for further analysis, interpretation, or improvement in computer vision, image processing, and related fields. It is a crucial step that involves manipulating the image data to enhance its quality, extract relevant information, or make it more suitable for our problem. These preprocessing steps as the following:
-
The initial step in preparing images for machine learning is resizing [63,64,65]. This process involves adjusting the image dimensions to a standardized size, promoting consistency throughout the dataset. By doing so, machine learning models make the images more easily processed. Additionally, resizing facilitates a reduction in computational complexity and memory requirements. However, selecting a size that optimizes efficiency while preserving essential image details is crucial.
-
The second step, called Color Transformation [66, 67], enables the conversion of images into a standardized color space, such as grayscale or RGB. This crucial step ensures consistency in color information across the dataset, facilitating models’ extraction of relevant features. Additionally, color transformation can enhance or simplify the visual information contained in the images.
-
In the third step, known as normalization [68, 69], pixel values are scaled to a standardized range, usually between 0 and 1. This process effectively eliminates any variations in intensity and contrast between images. By doing so, the model becomes less susceptible to discrepancies in illumination and color, ultimately enhancing its ability to generalize across varying lighting conditions.
-
During the final phase of image processing, labeling is performed to allocate categorical or numerical labels to every image, as demonstrated by [70, 71]. This step is pivotal for supervised machine learning endeavors since it facilitates the model’s ability to identify and associate visual characteristics with specific categories or results. Hence, precise labeling is essential for successful model training and assessment. In this paper, the data are labeled as 0 for MSIMUT and 1 for MSS and then shuffled and split as 80% training and 20% testing to be as in Table 1.
-
Quality Assessment Measures:
-
Image quality: We conducted a rigorous evaluation of the quality of histopathology images before preprocessing [72]. This assessment included checks for factors such as image resolution, clarity, and the presence of artifacts or distortions that might affect model performance.
-
Consistency check: The consistency of the dataset was evaluated by ensuring uniform labeling and normalization across all images [73, 74]. This step helps reduce bias and ensure the data’s robustness for deep learning analysis.
-
4.2 Working circumstances
The simulation has generated its results by utilizing powerful hardware, specifically an Intel Core i7 CPU, 64 GB of RAM, and an NVIDIA GTX 1050i GPU. Additionally, Python, Keras, Tensorflow, and Sklearn were utilized as programming tools to carry out the necessary programming tasks. Table 2 presents the recommended hyperparameters for the proposed models, while other standard parameter options, including the loss function and maximum number of epochs, are also available. The chosen optimizer for this task is Adam, and the loss function that was applied is shown in Table 2.
4.3 Evaluation metrics
In the field of predictive modeling, it is essential to assess the effectiveness of the presented models. Assessment measures such as recall (RC), precision (PR), accuracy, and F1 score (F1) are commonly used to evaluate the performance of these models. These metrics are determined by the assessment parameters known as true positive (TP), true negative (TN), false positive (FP), and false negative (FN). Accurate assessment measures are vital in determining the effectiveness of predictive models, and they can help businesses and academics make important decisions based on these models. Therefore, it is crucial to understand the significance of these metrics and how they relate to the assessment of predictive models.
The mathematical definition of recall (RC), as outlined in [75], is established based on the ratio of true positives (TP) to the product of TP and false negatives (FN). This relationship is depicted in Eq. 17.
To determine precision (PR), as described in [76], Eq. 18 is utilized. PR is calculated as TP divided by the product of TP and FP. This formula provides a quantitative measure of the accuracy of the results and helps assess the model’s effectiveness.
Equation 19 describes accuracy as follows:
The F1 score(F1) [77] measures a model’s accuracy by combining its PR and RC. It is calculated by multiplying the two values and dividing the result by their sum, and then multiplying by two. This formula is represented in Eq. 20.
Specificity (SP) is expressed as in Eq.(21):
Area under the curve (AUC) [78, 79] is a widely used metric in the field of ML for evaluating the performance of binary classification models. It is derived from the ROC (Receiver Operating Characteristic) curve, which is a graphical representation of a model’s ability to distinguish between two classes as the classification threshold varies. The AUC value ranges from 0 to 1, with higher values indicating better model performance. An AUC of 0.5 suggests random guessing, while an AUC of 1 signifies a perfect model.
4.4 Comparative analysis
This paper proposes a framework for the early detection of GI cancer, which faces the challenge of processing a large amount of data. Various DL models were employed to achieve high accuracy and efficiency in the shortest possible time to address this issue. A CNN-based model was proposed, followed by a hybrid model that combined CNNs with each type of RNN (SimpleRNN, LSTM, and GRU). A fully integrated model, CNN-SimpleRNN-LSTM-GRU, was developed to meet the objective.
4.4.1 The findings of the suggested CNNs
This section presents the simulation results of our suggested CNNs. Our models underwent 100 epochs with a batch size of 300, as outlined in Table 2. Figure 17 showcases the accuracy, loss curves, and confusion matrix of the CNNs. During the training phase, our suggested CNNs achieved an impressive accuracy of 98.53%. For the testing phase, we provide the evaluation metrics of the model in Table 3, which includes accuracy, PR, RC, SP, F1, and AUC, all measured at 97.10%, 95.6%, 92.4%, 98.6%, 94.0%, and 0.955, respectively.
4.4.2 The findings of the hybrid CNN-SimpleRNN model:
This section showcases the simulation results of the hybrid CNN-SimpleRNN model. Our model underwent training for 25 epochs with a batch size of 300, as outlined in Table 2. In Fig. 18, we present the model’s accuracy, loss curves, and confusion matrix. We are thrilled to announce that our hybrid model achieved a perfect 100.00% accuracy during training. Moving on to the testing phase, we evaluated the metrics of the CNN-SimpleRNN model, which we present in Table 4. Our model delivered impressive results, with accuracy, PR, RC, SP, F1, and AUC of 99.74%, 99.9%, 99.0%, 100.0%, 99.5%, and 0.995, respectively.
4.4.3 The findings of the hybrid CNN-LSTM model:
This section showcases the simulation results of the hybrid CNN-LSTM model. The model was trained using 65 epochs with a batch size of 350, as outlined in Table 2. Figure 19 illustrates the hybrid’s accuracy, loss curves, and confusion matrix. During training, the CNN-LSTM model achieved an accuracy of 100.00%. In the testing stage, the evaluation metrics are presented in Table 5, where the model achieved an accuracy, PR, RC, SP, F1, and AUC of 99.81%, 100.00%, 99.3%, 100.00%, 99.6%, and 0.996, respectively.
4.4.4 The findings of the hybrid CNN-GRU model:
Here, we present the simulation results of the hybrid CNN-GRU model. The model was trained with 80 epochs using a batch size of 300, as detailed in Table 2. Figure 20 showcases the hybrid model’s accuracy, loss curves, and confusion matrix. Notably, during training, the hybrid CNN-GRU model achieved an impressive accuracy of 99.75%. Moving onto the testing stage, we showcase the evaluation metrics of the hybrid CNN-GRU model in Table 6. Impressively, the model achieved exceptional accuracy, PR, RC, SP, F1, and AUC of 99.70%, 99.8%, 99.0%, 99.9%, 99.4%, and 0.995, respectively.
4.4.5 The findings of the hybrid CNN-SimpleRNN-LSTM-GRU model
In this section, we present the simulation results of the hybrid CNN-SimpleRNN-LSTM-GRU model. Our model underwent 100 training epochs with a batch size of 350, as outlined in Table 2. As depicted in Fig. 21, the accuracy, loss curves, and confusion matrix are displayed for our hybrid model. During training, our hybrid model achieved a perfect accuracy of 100.00%. We evaluated our hybrid model using various metrics in Table 7 in the testing phase. Our model achieved impressive scores for accuracy, PR, RC, SP, F1, and AUC, with results of 99.90%, 99.9%, 99.6%, 100.00%, 99.8%, and 1.00, respectively.
4.4.6 Results discussion
After careful analysis, it is observed that the CNNS-SimpleRNN-LSTM-GRU hybrid model is the top performer. It boasts outstanding real-time performance and outperforms all other models in nearly every metric. This model is a true all-rounder, ranking first in accuracy, RC, F1, SP, AUC, loss, and processing time, with a respectable second-place ranking in PR. The CNNs-LSTM hybrid is also a strong contender, taking first place in PR and SP and second place in accuracy, RC, F1, AUC, and processing time, with a third-place ranking in loss. The CNNs-SimpleRNN hybrid is a close third, ranking first in SP, second in PR and loss, and third in accuracy, RC, F1, and AUC, with a fourth-place ranking in processing time. The CNNs-GRU hybrid is also a worthy competitor, securing second place in SP and third place in PR, RC, AUC, and processing time, with a fourth-place ranking in accuracy, F1, and loss. Regrettably, the Suggested CNNs model did not meet the standard and ranked last in all measures (Table 8).
It has also been proved that the combination of CNN and RNN families has shown promising results, especially when considering time as a significant factor. Overcoming the challenge posed by time was a critical aspect of our approach, as we aimed to achieve the most optimal outcomes. The results obtained demonstrated the remarkable success of our methodology. It is confirmed more in Sect. 4.5.
These findings have significant clinical implications. High accuracy and recall are crucial for correctly identifying cancerous patterns, reducing false negatives, and ensuring timely intervention. Specificity ensures that healthy tissues are not misclassified, minimizing unnecessary treatments. The high AUC values indicate robust model performance across different thresholds, enhancing reliability in clinical settings as the following:
-
High specificity and sensitivity: The model’s high specificity (SP of 100.0) ensures that it correctly identifies patients without the disease, reducing false positives. This minimizes unnecessary treatments and anxiety for patients. The high sensitivity (RC of 99.6) ensures that most cases of the disease are correctly identified, which is crucial for early detection and treatment.
-
Early detection: Due to its high accuracy and swift processing times, this hybrid model can be seamlessly integrated into clinical workflows to support the early detection of gastrointestinal cancer. Early diagnosis significantly improves patient outcomes by enabling timely intervention.
-
Reduction in diagnostic time: The model’s efficient processing times (training time of 8 min and testing time of 3 s) enables rapid analysis of patient data, offering quick diagnostic results. This efficiency is particularly valuable in clinical settings where prompt decision-making is essential.
-
Potential for personalized medicine: The model’s precise classification capabilities can aid in the development of personalized treatment plans. By accurately identifying MSI and MSS patterns, clinicians can customize treatment strategies based on the specific genetic and molecular characteristics of the tumor.
4.5 Comparison with the state-of-the-art models
According to the findings in Table9, the hybrid of CNNs-LSTM effectively improves precision measures. However, the hybrid of CNNs-SimpleRNN-LSTM-GRU surpasses the other proposed models in almost all measures, including processing time. Notably, this hybrid model also outperforms similar models using the same dataset. This can be attributed to its various advantages, which include:
-
Spatial and temporal features integration: A hybrid model incorporating CNN for spatial feature extraction and RNN layers (SimpleRNN, LSTM, and GRU) for sequential or temporal information processing allows for the integration of both spatial and temporal features in a single model. This can be crucial in tasks where both types of information are relevant, such as video analysis.
-
Efficient information capture: CNN layers excel at capturing spatial information from images, while RNN layers are designed to capture sequential patterns and dependencies. This combination enables the model to efficiently capture and exploit information across different dimensions in the data.
-
Sequential context handling: RNN layers (SimpleRNN, LSTM, and GRU) are well suited for tasks that require an understanding of sequential context, like recognizing patterns or temporal dependencies within image sequences, making them beneficial for various scientific applications.
-
Reduced overfitting: Including RNN layers can help mitigate overfitting, as they can capture long-range dependencies and smooth out noise in the data. This can result in more robust and generalized models, especially in cases where data are limited.
-
Adaptive model learning: The dynamic nature of RNN layers, such as LSTM and GRU, allows the model to adapt to varying sequences or temporal patterns, enhancing the model’s ability to handle different types of input data.
-
Contextual image understanding: A hybrid model can better understand the contextual relationships within images for scientific image analysis. It can capture not only individual object features but also how they relate to each other over time or across different image regions.
-
Complex pattern recognition: The combination of CNN for feature extraction and RNN for sequential analysis is powerful for recognizing complex image patterns. This can be valuable in scientific domains where identifying intricate structures or phenomena is crucial.
-
Interpretability and explainability: Hybrid models are often more interpretable as they allow you to analyze spatial and temporal features separately. This can be advantageous in scientific research, where understanding the model’s decision-making process is essential.
-
Customization for specific tasks: The hybrid architecture can be customized to suit the unique requirements of your image classification task, making it highly adaptable and efficient for scientific applications that may have specialized needs.
-
Scientific insights: By combining these neural network components in a hybrid model, you may gain scientific insights into the interactions between spatial and temporal information in your data, contributing to a deeper understanding of the underlying phenomena.
In addition to the advantages mentioned earlier, a hybrid CNN-SimpleRNN-LSTM-GRU model proved its efficiency in decreasing processing time because of the following:
-
Parallel processing: CNN layers are well suited for parallel processing, as they can simultaneously analyze different regions of an image. This parallelism can lead to faster feature extraction compared to sequential processing.
-
Reduced computation: RNN layers, especially LSTM and GRU, are computationally more efficient than fully connected layers. By incorporating these layers, you can reduce the overall computational burden, leading to faster inference times.
-
Hierarchical feature extraction: The combination of CNN for spatial feature extraction and RNN for temporal processing enables a hierarchical approach, where spatial features are extracted first, and then, the sequential analysis is performed. This can lead to more efficient utilization of computational resources.
-
Early exit mechanisms: Hybrid models can be designed with early exit mechanisms, allowing the model to make predictions at intermediate stages. This can be useful in real-time applications, where quick decisions based on partial information may be needed, saving processing time.
-
Reduced model complexity: A well-designed hybrid model can be more compact than a standalone deep CNN or RNN. Reduced model complexity leads to faster training and inference times, making it suitable for applications with strict time constraints.
-
Efficient memory usage: RNN layers, especially GRU, have a more efficient memory management mechanism than standard RNNs. This can help reduce memory consumption, which is crucial for processing large images or long sequences.
-
Optimized hyperparameters: The flexibility of the hybrid architecture allows you to optimize hyperparameters for faster training and inference. To balance accuracy and speed, you can experiment with layer sizes, batch sizes, and learning rates.
-
Effective utilization of hardware: Modern hardware, such as GPUs and TPUs, is designed to handle various operations efficiently. By combining CNN and RNN layers, you can better utilize the capabilities of these hardware accelerators for faster computations.
-
Model pruning: Hybrid models can be pruned to remove unnecessary connections or layers, resulting in a more streamlined model. Pruned models are faster to execute, which is essential for real-time applications.
-
Streaming data processing: In scenarios where you need to process streaming or real-time data, hybrid models can be more responsive due to their efficient spatial and temporal processing combination, enabling faster decision-making.
The findings in Table 9 also corroborate the clinical relevance and effectiveness of hybrid deep learning models, emphasizing their significant role in medical diagnostics as the following:
-
Integration of spatial and temporal features: Studies by Yamashita et al. [24] and Kather et al. [22] demonstrate the enhanced model performance when combining spatial features captured by CNNs with temporal features processed by RNNs, consistent with our results.
-
Improved performance metrics: Our high-performance metrics align with the results reported by Chen et al. [23] and Qiu et al. [32], indicating that hybrid models surpass traditional methods in accuracy and diagnostic capability.
-
Clinical relevance: Research conducted by Khan et al. [28] and Lee et al. [29] underscores the clinical importance of deep learning models in cancer diagnosis. Our study reinforces these findings, highlighting the practical utility of a hybrid CNN-SimpleRNN-LSTM-GRU model in clinical environments.
-
Efficiency and processing time: The efficient processing times we observe align with findings from Zhu et al. [30].
5 Conclusion
Our investigation into the early detection of GI cancer through The integration of histopathology data, coupled with The application of five distinct DL models, has yielded valuable insights. Notably, our findings emphasize the pivotal role of MSI and MSS in refining diagnostic accuracy. Among the models evaluated, the CNN-SimpleRNN-LSTM-GRU architecture emerged as the frontrunner, excelling across critical metrics such as accuracy, RC, SP, F1, and AUC. This model’s nuanced integration of recurrent layers showcases its efficacy in capturing temporal dependencies within the histopathology data, thus enhancing its discriminatory power in distinguishing between MSI and MSS cases. However, it is imperative to recognize each model’s inherent advantages and limitations. While the CNN-LSTM demonstrated commendable PR, the computational demands of more intricate architectures warrant careful consideration, especially in resource-constrained settings. Furthermore, the varying performance across models underscores the importance of tailoring diagnostic approaches to specific clinical requirements, considering the distinct characteristics of MSI and MSS patterns. Future research endeavors could delve deeper into unraveling the intricate molecular signatures associated with MSI and MSS, refining the specificity of diagnostic models. Exploring the transferability of our findings across diverse patient cohorts and datasets would contribute to the robustness of the proposed diagnostic framework. Additionally, investigations into the interpretability of these DL models concerning the nuanced histopathological features linked to MSI and MSS could facilitate greater clinical acceptance. In conclusion, our study not only underscores the promising potential of DL in early GI cancer detection but also highlights the critical role of MSI and MSS stratification in enhancing diagnostic precision. The ongoing pursuit of optimized models, informed by a comprehensive understanding of molecular intricacies, is essential for advancing the field and realizing the full clinical impact of AI-driven diagnostics in oncology.
Data availability
Data are available on request from the authors.
Abbreviations
- DL:
-
Deep learning
- RNNs:
-
Recurrent neural networks
- LSTM:
-
Long short-term memory
- MSI:
-
Microsatellite instability
- TCGA:
-
The Cancer Genome Atlas
- GC:
-
Gastric cancer
- WSIs:
-
Whole slide images
- ANNs:
-
Artificial neural networks
- SVM:
-
Support vector machines
- DLCT:
-
Dual-layer computed tomography
- DNN:
-
Deep neural network
- TP:
-
True positive
- FP:
-
False positive
- BPTT:
-
Backpropagation through time
- PR:
-
Precision
- SP:
-
Specificity
- ML:
-
Machine learning
- CNNs:
-
Convolutional neural networks
- GRU:
-
Gated recurrent unit
- MSS:
-
Microsatellite stability
- GI:
-
Gastrointestinal
- CRC:
-
Colorectal cancer
- MMR:
-
Mismatch repair
- CT:
-
Computer tomography
- NPV:
-
Net present value
- SL:
-
Swarm learning
- ReLU:
-
Rectified linear unit
- TN:
-
True negative
- FN:
-
False negative
- AUC:
-
Area under the curve
- RC:
-
Recall
- F1:
-
F1 score
References
Xie Y, Shi L, He X, Luo Y (2021) Gastrointestinal cancers in China, the USA, and Europe. Gastroenterol Rep 9(2):91–104
Ağagündüz D, Cocozza E, Cemali Ö, Bayazıt AD, Nanì MF, Cerqua I, Morgillo F, Saygılı SK, Berni Canani R, Amero P et al (2023) Understanding the role of the gut microbiome in gastrointestinal cancer: a review. Front Pharmacol 14:1130562
Wang J, Xiu J, Farrell A, Baca Y, Arai H, Battaglin F, Kawanishi N, Soni S, Zhang W, Millstein J et al (2023) Mutational analysis of microsatellite-stable gastrointestinal cancer with high tumour mutational burden: a retrospective cohort study. Lancet Oncol 24(2):151–161
Yamashita K, Arimura Y, Kurokawa S, Itoh F, Endo T, Hirata K, Imamura A, Kondo M, Sato T, Imai K (2000) Microsatellite instability in patients with multiple primary cancers of the gastrointestinal tract. Gut 46(6):790–794
Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T (2017) Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget 8(67):112103
Shinozaki-Ushiku A, Kunita A, Iwasaki A, Kato M, Yamazawa S, Abe H, Ushiku T (2023) Microsatellite instability profiles of gastrointestinal cancers: comparison between non-colorectal and colorectal origin. Histopathology 82(3):466–477
Marrelli D, Polom K, Pascale V, Vindigni C, Piagnerelli R, De Franco L, Ferrara F, Roviello G, Garosi L, Petrioli R et al (2016) Strong prognostic value of microsatellite instability in intestinal type non-cardia gastric cancer. Ann Surg Oncol 23:943–950
Zito Marino F, Amato M, Ronchi A, Panarese I, Ferraraccio F, De Vita F, Tirino G, Martinelli E, Troiani T, Facchini G et al (2022) Microsatellite status detection in gastrointestinal cancers: PCR/NGS is mandatory in negative/patchy MMR immunohistochemistry. Cancers 14(9):2204
Villalobos P, Wistuba II (2017) Lung cancer biomarkers, hematology/oncology. Clinics 31(1):13–29
Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN (2021) Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer 124(4):686–696
Khan MA, Sahar N, Khan WZ, Alhaisoni M, Tariq U, Zayyan MH, Kim YJ, Chang B (2022) Gestronet: A framework of saliency estimation and optimal deep learning features based gastrointestinal diseases detection and classification. Diagnostics 12(11):2718
Kim SY, Kim TW (2020) Current challenges in the implementation of precision oncology for the management of metastatic colorectal cancer. ESMO Open 5(2):e000634
Magalhães G, Calisto R, Freire C, Silva R, Montezuma D, Canberk S, Schmitt F (2024) Invisible for a few but essential for many: the role of histotechnologists in the establishment of digital pathology. J Histotechnol 47(1):39–52
Ben-Aharon I, van Laarhoven HW, Fontana E, Obermannova R, Nilsson M, Lordick F (2023) Early-onset cancer in the gastrointestinal tract is on the rise-evidence and implications. Cancer Discov 13(3):538–551
Ahmed IA, Senan EM, Shatnawi HSA (2023) Hybrid models for endoscopy image analysis for early detection of gastrointestinal diseases based on fused features. Diagnostics 13(10):1758
Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM, Giger ML (2019) Deep learning in medical imaging and radiation therapy. Med Phys 46(1):e1–e36
Shafi S, Parwani AV (2023) Artificial intelligence in diagnostic pathology. Diagn Pathol 18(1):109
Jiang Y, Yang M, Wang S, Li X, Sun Y (2020) Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun 40(4):154–166
Huss R, Raffler J, Märkl B (2023) Artificial intelligence and digital biomarker in precision pathology guiding immune therapy selection and precision oncology. Cancer Rep 6:e1796
Min JK, Kwak MS, Cha JM (2019) Overview of deep learning in gastrointestinal endoscopy. Gut Liver 13(4):388
Jaspers TJ, Boers, TG, Kusters CH, Jong MR, Jukema JB, de Groof AJ, Bergman JJ, de With PH, van der Sommen F (2023) Investigating the impact of image quality on endoscopic ai model performance. In: International workshop on applications of medical AI, Springer, pp 32–41
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP et al (2019) Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25(7):1054–1056
Chen T, Zhang C, Liu Y, Zhao Y, Lin D, Hu Y, Yu J, Li G (2019) A gastric cancer LncRNAs model for MSI and survival prediction based on support vector machine. BMC Genomics 20(1):1–7
Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, Higgins J, Rubin DL, Shen J (2021) Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol 22(1):132–141
Cao R, Yang F, Ma S-C, Liu L, Zhao Y, Li Y, Wu D-H, Wang T, Lu W-J, Cai W-J et al (2020) Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in colorectal cancer. Theranostics 10(24):11080
Zhang Z, Peng J (2023) Clinical nursing and postoperative prediction of gastrointestinal cancer based on CT deep learning model. J Radiat Res Appl Sci 16(2):100561
Padmavathi P, Harikiran J (2023) Se-resnet: A novel method for gastrointestinal (GI) diseases classification from wireless capsule endoscopy (WCE) images. Traitement du Signal 40(4)
Khan Z, Loganathan R (2022) Transfer Learning Based Classification of MSI and MSS Gastrointestinal Cancer. EasyChair
Lee SH, Lee Y, Jang H-J (2023) Deep learning captures selective features for discrimination of microsatellite instability from pathologic tissue slides of gastric cancer. Int J Cancer 152(2):298–307
Zhu Y, Wang P, Wang B, Jiang Z, Li Y, Jiang J, Zhong Y, Xue L, Jiang L (2023) Dual-layer spectral-detector CT for predicting microsatellite instability status and prognosis in locally advanced gastric cancer. Insights Imaging 14(1):151
Saldanha OL, Muti HS, Grabsch HI, Langer R, Dislich B, Kohlruss M, Keller G, van Treeck M, Hewitt KJ, Kolbinger FR et al (2023) Direct prediction of genetic aberrations from pathology images in gastric cancer with swarm learning. Gastric Cancer 26(2):264–274
Qiu W, Yang J, Wang B, Yang M, Tian G, Wang P, Yang J (2022) Evaluating the microsatellite instability of colorectal cancer based on multimodal deep learning integrating histopathological and molecular data. Front Oncol 12:925079
Yu S-N, Huang S-C, Wang W-C, Chang YP, Chen K-H, Chen T-C (2023) Prediction of microsatellite instability from gastric histological images based on residual attention networks with non-local modules, IEEE Access
Yin H, Vahdat A, Alvarez JM, Mallya A, Kautz J, Molchanov P (2022) A-vit: adaptive tokens for efficient vision transformer. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 10809–10818
Ren K, Hong G, Chen X, Wang Z (2023) A covid-19 medical image classification algorithm based on transformer. Sci Rep 13(1):5359
Azad R, Kazerouni A, Heidari M, Aghdam EK, Molaei A, Jia Y, Jose A, Roy R, Merhof D (2023) Advances in medical image analysis with vision transformers: a comprehensive review. Med Image Anal 91:103000
Meng L, Li H, Chen B-C, Lan S, Wu Z, Jiang Y-G, Lim S-N (2022) Adavit: adaptive vision transformers for efficient image recognition. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 12309–12318
Yin H, Vahdat A, Alvarez J, Mallya A, Kautz J, Molchanov P (2021) Adavit: adaptive tokens for efficient vision transformer, arXiv preprint arXiv:2112.07658
Chen C-FR, Fan Q, Panda R (2021) CrossViT: Cross-attention multi-scale vision transformer for image classification, in: Proceedings of the IEEE/CVF international conference on computer vision, pp 357–366
Xu H, Zheng T, Liu Y, Zhang Z, Xue C, Li J (2024) A joint convolutional cross vit network for hyperspectral and light detection and ranging fusion classification. Remote Sens 16(3):489
Zheng Y, Jiang W (2022) Evaluation of vision transformers for traffic sign classification. Wirel Commun Mob Comput 2022(1):3041117
Tummala S, Kadry S, Bukhari SAC, Rauf HT (2022) Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Curr Oncol 29(10):7498–7511
Chauhan R, Ghanshala KK, Joshi R (2018) Convolutional neural network (CNN) for image detection and recognition. In: First international conference on secure cyber computing and communication (ICSCCC). IEEE, pp 278–282
Albawi S, Mohammed TA, Al-Zawi S (2017) Understanding of a convolutional neural network. In: International conference on engineering and technology (ICET). IEEE, pp 1–6
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging 9:611–629
O’Shea K, Nash R (2015) An introduction to convolutional neural networks, arXiv preprint arXiv:1511.08458
Dhruv P, Naskar S (2019) Image classification using convolutional neural network (CNN) and recurrent neural network (RNN): a review. Machine Learning Inf Processing Proc ICMLIP 2020:367–381
Grossberg S (2013) Recurrent neural networks. Scholarpedia 8(2):1888
Medsker LR, Jain L (2001) Recurrent neural networks. Design Appl 5(64–67):2
Salehinejad H, Sankar S, Barfett J, Colak E, Valaee S (2017) Recent advances in recurrent neural networks, arXiv preprint arXiv:1801.01078
Ma Y, Principe JC (2019) A taxonomy for neural memory networks. IEEE Trans Neural Netw Learn Syst 31(6):1780–1793
Kag A, Saligrama V (2021) Time adaptive recurrent neural network. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 15149–15158
Yu Y, Si X, Hu C, Zhang J (2019) A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput 31(7):1235–1270
Staudemeyer RC, Morris ER (2019) Understanding LST–a tutorial into long short-term memory recurrent neural networks, arXiv preprint arXiv:1909.09586
Smagulova K, James AP (2019) A survey on lSTM memristive neural network architectures and applications. Eur Phy J Spec Top 228(10):2313–2324
Sherstinsky A (2020) Fundamentals of recurrent neural network (RNN) and long short-term memory (ISTM) network. Phys D 404:132306
Dey R, Salem FM (2017) Gate-variants of gated recurrent unit (GRU) neural networks. In: IEEE 60th international Midwest symposium on circuits and systems (MWSCAS). IEEE, pp 1597–1600
Shewalkar A, Nyavanandi D, Ludwig SA (2019) Performance evaluation of deep neural networks applied to speech recognition: RNN, LSTM and GRU. J Artif Intell Soft Comput Res 9(4):235–245
Fu R, Zhang Z, Li L (2016) Using LSTM and GRU neural network methods for traffic flow prediction. In: 31st Youth academic annual conference of Chinese association of automation (YAC). IEEE, pp 324–328
Awcock GJ, Thomas R (1995) Applied image processing. Springer
Petrou MM, Petrou C (2010) Image processing: the fundamentals. John Wiley & Sons
Huang TS, Schreiber WF, Tretiak OJ (1971) Image processing. Proc IEEE 59(11):1586–1609
Sullivan BJ, Ansari R, Giger ML, MacMahon H (1995) Effects of image preprocessing/resizing on diagnostic quality of compressed medical images [chest radiographs application]. In: Proceedings., international conference on image processing, Vol 2, IEEE, pp 13–16
Perumal S, Velmurugan T (2018) Preprocessing by contrast enhancement techniques for medical images. Int J Pure Appl Math 118(18):3681–3688
Brisinello M, Grbić R, Pul M, Anđelić T (2017) Improving optical character recognition performance for low quality images. In: International symposium ELMAR. IEEE, pp 167–171
Förstner W (2003) Image preprocessing for feature extraction in digital intensity, color and range images, in: Geomatic method for the analysis of data in the earth sciences, Springer, pp 165–189
Khirade SD, Patil A (2015) Plant disease detection using image processing. In: International conference on computing communication control and automation. IEEE, pp 768–771
Sharif M, Mohsin S, Jamal MJ, Raza M (2010) Illumination normalization preprocessing for face recognition. In: The 2nd conference on environmental science and information application technology, vol 2. IEEE, pp 44–47
Meier J, Bock R, Michelson G, Nyúl LG, Hornegger J (2007) Effects of preprocessing eye fundus images on appearance based glaucoma classification. In: Computer analysis of images and patterns: 12th international conference, CAIP 2007, Vienna, Austria, August 27–29, 2007. Proceedings 12, Springer, pp 165–172
Caseneuve G, Valova I, LeBlanc N, Thibodeau M (2021) Chest X-ray image preprocessing for disease classification. Procedia Comput Sci 192:658–665
Pérez-García F, Sparks R, Ourselin S (2021) Torchio: a python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Comput Methods Programs Biomed 208:106236
Shiranita K, Hayashi K, Otsubo A, Miyajima T, Takiyama R (2000) Grading meat quality by image processing. Pattern Recogn 33(1):97–104
Amato G, Falchi F, Gennaro C (2011) Geometric consistency checks for kNN based image classification relying on local features. In: Proceedings of the fourth international conference on similarity search and applications, pp 81–88
S. Sergyán, L. Csink (2005) Consistency check of image databases. In: Proceedings of 2nd Romanian-Hungarian joint symposium on applied computational intelligence, Timisoara, Romania, Citeseer, pp 201–206
Amigó E, Gonzalo J, Artiles J, Verdejo F (2009) A comparison of extrinsic clustering evaluation metrics based on formal constraints. Inf Retr 12:461–486
De Medeiros AKA, Guzzo A, Greco G, Van der Aalst WM, Weijters A, Van Dongen BF, Saccà D (2008) Process mining based on clustering: a quest for precision. In: Business process management workshops: BPM 2007 international workshops, BPI, BPD, CBP, ProHealth, RefMod, semantics4ws, Brisbane, Australia, September 24, 2007, Revised Selected Papers 5, Springer, pp 17–29
Amigó E, Gonzalo J, Artiles J, Verdejo F (2011) Combining evaluation metrics via the unanimous improvement ratio and its application to clustering tasks. J Artif Intell Res 42:689–718
Lobo JM, Jiménez-Valverde A, Real R (2008) AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr 17(2):145–151
Huang J, Ling CX (2005) Using AUC and accuracy in evaluating learning algorithms. IEEE Trans Knowl Data Eng 17(3):299–310
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wafa, A.A., Essa, R.M., Abohany, A.A. et al. Integrating deep learning for accurate gastrointestinal cancer classification: a comprehensive analysis of MSI and MSS patterns using histopathology data. Neural Comput & Applic (2024). https://doi.org/10.1007/s00521-024-10287-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00521-024-10287-y