Abstract
Nowadays, machine learning (ML) has attained a high level of achievement in many contexts. Considering the significance of ML in medical and bioinformatics owing to its accuracy, many investigators discussed multiple solutions for developing the function of medical and bioinformatics challenges using deep learning (DL) techniques. The importance of DL in Internet of Things (IoT)-based bio- and medical informatics lies in its ability to analyze and interpret large amounts of complex and diverse data in real time, providing insights that can improve healthcare outcomes and increase efficiency in the healthcare industry. Several applications of DL in IoT-based bio- and medical informatics include diagnosis, treatment recommendation, clinical decision support, image analysis, wearable monitoring, and drug discovery. The review aims to comprehensively evaluate and synthesize the existing body of the literature on applying deep learning in the intersection of the IoT with bio- and medical informatics. In this paper, we categorized the most cutting-edge DL solutions for medical and bioinformatics issues into five categories based on the DL technique utilized: convolutional neural network, recurrent neural network, generative adversarial network, multilayer perception, and hybrid methods. A systematic literature review was applied to study each one in terms of effective properties, like the main idea, benefits, drawbacks, methods, simulation environment, and datasets. After that, cutting-edge research on DL approaches and applications for bioinformatics concerns was emphasized. In addition, several challenges that contributed to DL implementation for medical and bioinformatics have been addressed, which are predicted to motivate more studies to develop medical and bioinformatics research progressively. According to the findings, most articles are evaluated using features like accuracy, sensitivity, specificity, F-score, latency, adaptability, and scalability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Bioinformatics synthesizes computer programming, biology, and big data to aid scientists in perceiving and detecting paradigms in biological and medical information [1,2,3]. It is significantly suitable for studying DNA sequencing, as it allows scientists to arrange a great deal of data [4, 5]. The area of computer science, namely bioinformatics, is applied to evaluate whole-genome sequencing information [6, 7]. This contains software improvement, algorithm, analysis, pipeline, transferring, and storage/database improvement of genomics information. In other words, bioinformatics is described as applying analysis and computation tools to receive and interpret biological data [8, 9]. As an interdisciplinary area, bioinformatics harnesses computer science, physics, biology, and mathematics [10, 11]. It is critical for data management in modern medicine and biology [12, 13]. Bioinformatics provides considerable support to deal with time context and cost issues in different tracks [14, 15]. Bioinformatics, as pertinent to genomics and genetics, is a scientific interdisciplinary field that utilizes computer technology to gather, store, evaluate, and distribute biological data, like DNA and amino acid sequences and annotations about them [16, 17].
Distributed computing is a versatile method that can be applied to a wide range of issues in bioinformatics. While it is commonly used for cost-efficiency in high-performance computing, in other domains, it has become a necessity [18]. By leveraging the power of multiple interconnected computers, distributed computing allows researchers to process large amounts of data and perform complex calculations quickly and efficiently. This is particularly important in bioinformatics, where massive data sets are often analyzed to gain insights into biological processes and develop new disease treatments [19, 20]. The rising amount of information required to be processed within logical time finally outgrows even the strongest computers [21, 22]. Recently, some organizations that regularly process large amounts of data have yet to apply nearly easy processes, like distributing work by hand or with easy scripts [23, 24]. An increasing number of corporations are suggesting solutions that scale better, allowing more autonomy of information analysis procedures and more effective resource usage [25, 26]. Some present tools for distributed computing are too low-phase or not flexible enough to be adjusted to requirements [27, 28]. Many technologies generate a great base for building higher-level distributed computing ecosystems [29, 30]. Also, Machine learning (ML), a subsection of Artificial Intelligence (AI), has become a strong tool for several bioinformatics uses [31, 32]. Depending on big datasets, ML mechanisms are particularly suitable for forecasting and pattern recognition [33]. There are some emerging uses of ML within the bioinformatics area. ML in bioinformatics refers to using ML techniques to analyze and interpret biological data, including genomics, systems biology, text mining, microarrays, and evolution. By applying ML algorithms to complex biological data sets, researchers can gain insights into various biological processes, identify genetic mutations, and even develop new disease treatments [34, 35]. ML can be applied through various modes of human-made database reports to process and evaluate data, decreasing labor expenses and fastening the research procedure without compromising quality [36, 37]. ML text evaluation can be utilized in bioinformatics as well. The containing of ML has given bioinformatics the needed promotion [38, 39].
This research aimed to provide a detailed overview of the applications of ML technologies in IoT-based medical and bioinformatics. The study highlighted the multiple uses of Deep Learning (DL) strategies [40, 41] in medical and bioinformatics by conducting an SLR and analyzing and comparing findings from various studies. The DL mechanisms used in medicine and bioinformatics were divided into five separate groups: convolutional neural network (CNN), recurrent neural network (RNN), generative adversarial network (GAN), multilayer perception (MLP), and hybrid approaches, which include several practical methods. For each group and mechanism, multiple properties such as benefits, drawbacks, datasets, and simulation environments were studied. The study investigated the methodologies and applications of DL/ML mechanisms in bioinformatics before delving further into future studies and taking into account the shortcomings that need to be addressed in the future. Overall, the contributions of this paper include providing a thorough examination of current concerns with ML/DL mechanisms in medical and bioinformatics, conducting a comprehensive evaluation of existing methods for ML/DL applications, and modeling important areas for the future development of these approaches. The main contributions of this paper are as follows:
-
Conducting an SLR to explore ML applications in IoT-based medical and bioinformatics;
-
Analyzing and comparing DL uses in medical and bioinformatics;
-
Categorizing DL mechanisms into five groups (CNN, RNN, GAN, MLP, hybrids) and examining their properties;
-
Investigating DL/ML methodologies and applications in bioinformatics;
-
Providing insights for future research and addressing existing shortcomings;
-
Offering a comprehensive evaluation of existing ML/DL methods;
-
Contributing to a better understanding of current challenges and opportunities in the field;
The article is structured in the following manner: The key principles and terminology of ML/DL in medical and bioinformatics are covered in the first part, followed by an investigation of relevant papers in part 3. Part 4 discusses the studied mechanisms and tools for paper selection, while Part 5 illustrates the classification that was selected. Section 6 presents the results and comparisons; Sect. 7 provides the open issues, and the conclusion is explored in Sect. 8.
2 Fundamental concepts and terminology
This section discusses the fundamentals of DL approaches as well as their applications in medical and bioinformatics.
2.1 Deep learning concepts
Three classifications of DL methods are supervised, semi-supervised, and unsupervised learning. An input vector as a value for the supervisory signal is a desired value. Present labels aid the method of predicting the desired output labels [42]. Classification approaches employ supervised learning to detect faces and traffic signals, translate voice to text, identify spam in a file, and perform a variety of other tasks. Semi-supervised learning is a strategy that crosses the gap between unsupervised and supervised ML approaches [43]. This approach, which falls between supervised and unsupervised learning, uses unlabeled and labeled values as training data. When combined with a modest quantity of labeled data, the learning accuracy of unlabeled data improves significantly. In theory, the data adjacent to it have the same name. Likewise, the cluster assumption, which states that every data in a cluster is the same, has a similar name [44]. Also, rather than using the whole input space, the data are limited to a single dimension. Unsupervised learning describes the interrelationships between the components and then categorizes them. These algorithms are used in neural networks, clustering, and anomaly detection. Detecting anomalies typically takes the benefit of unsupervised learning, specifically in security areas. By the same token, feature processing and extraction are possible by using DL techniques and artificial neural networks [45, 46].
2.2 Bioinformatics applications
Bioinformatics is an advanced field of biology that proceeds from the combination of both information and biology [47]. It is an interdisciplinary area of study that utilizes mathematics, biology, computer science, chemistry, and statistics, which have been synthesized to shape an individual order [48]. Bioinformatics is fundamentally applied to bring out knowledge from biological data through the improvement of software and algorithms [49]. Bioinformatics is broadly used in the study of Genomics, 3D structure modeling of Proteins, proteomics, image analysis, and drug designing [50, 51]. A particular use of bioinformatics can be found in the domain of preventive medicine, which is principally concentrated on improving measures to avoid, manage, and treat serious infectious diseases [52]. The basic target of bioinformatics is to enhance the understanding of biological procedures. There are several applications of bioinformatics including recording and retrieval of data in gene therapy, biometrical evaluation for crop management, pest control, evolutionary research, drug discovery, and microbial utilitarianism [52].
2.3 Deep learning usage in bioinformatics
The primary objective of healthcare informatics is to offer better treatments and enhance the quality of life for individuals by efficiently analyzing biomedical data, which includes Electronic Health Records (EHRs) [53]. In the past, it was customary to rely on domain experts to develop models for healthcare or biomedicine, but recent advances in DL algorithms have enabled the automatic learning of representations and patterns from such data for model improvement. DL techniques involve several levels of representation, where at each stage, the system learns higher abstract representations. Natural language processing (NLP), computer vision, speech recognition, video analysis, health informatics, and image processing are among the fields in which DL-based algorithms have performed well. Powerful computational models include DL approaches such as CNN, neural networks, auto-encoders, and deep generative networks. These techniques have shown considerable success in dealing with large amounts of information across a wide range of applications due to their ability to extract complex latent features and learn effective representations in an unsupervised setting [54]. Here are several uses of DL methods in bioinformatics medical systems:
2.3.1 Detecting enzymes applying multilayer neural networks
Detecting enzymes using multilayer neural networks refers to using DL algorithms to automatically recognize enzymes in biochemical data. Enzymes are proteins that catalyze chemical reactions in living organisms. Detecting and identifying enzymes is crucial in many areas of bioinformatics and biomedicine, such as drug discovery and metabolic pathway analysis. Traditionally, this task required the expertise of domain specialists to identify enzymes manually [55]. With the advancement of DL algorithms, it is now possible to train multilayer neural networks to recognize patterns in enzyme data and classify them automatically. Multilayer neural networks are a type of AI that consists of multiple layers of interconnected nodes that process input data to generate output predictions. These networks can learn to represent complex relationships between input features and output classes, making them effective for enzyme detection. The paper discusses various approaches to applying multilayer neural networks for enzyme detection, including the use of CNN and RNN. These networks can be trained on large datasets of enzyme data, and the resulting models can be used to automatically detect and classify enzymes in new data [56].
2.3.2 Gene expression regression
Gene expression regression refers to the use of DL algorithms to predict the expression level of a gene based on various factors, such as environmental conditions, genetic mutations, or other molecular processes. The goal is to build a model that can accurately predict the level of gene expression in a particular context, which can help researchers understand the underlying biological mechanisms and develop new treatments for diseases [57]. DL models, such as CNN or RNN, are trained on large datasets of gene expression data, along with other relevant features. These models learn to identify patterns and correlations between the expression levels and the various factors that influence them, allowing them to make accurate predictions. Gene expression regression has numerous applications in bioinformatics and medical informatics, including predicting drug responses, identifying biomarkers for diseases, and understanding the mechanisms of genetic disorders [58].
2.3.3 CNN predicting RNA–protein linking points
In bioinformatics, predicting RNA–protein binding sites is an important task as it can help in understanding gene regulation, disease diagnosis, and drug discovery. One approach to this task is using CNN [59], which is a type of DL model designed to learn spatial features from input data. In the context of predicting RNA–protein binding sites, CNNs can be trained on sequence data to identify patterns and features that are indicative of RNA–protein interaction sites [60]. The input to the CNN is a sequence of nucleotides, and the output is a probability score that indicates the likelihood of RNA–protein binding at each position in the sequence. CNN works by applying a set of filters to the input sequence, with each filter looking for a specific pattern or feature in the sequence. The output of the filter is then passed through a nonlinear activation function to generate a feature map. Multiple filters are used in parallel to learn different features from the input sequence. The feature maps are then pooled to reduce the dimensionality of the data and to capture the most salient features. The resulting features are then passed through one or more fully connected layers to make the final prediction. Overall, the use of CNNs for predicting RNA–protein binding sites has shown promising results and has the potential to contribute to developing new therapeutics and diagnostics for various diseases [61].
2.3.4 DNA sequence performance anticipation with RNN and CNN
DNA sequence performance anticipation with RNN and CNN refers to the use of RNN and CNN in predicting the performance of DNA sequences. RNNs are neural networks designed to process sequential data by maintaining a memory of past inputs, while CNNs are a type of neural network that can learn and identify spatial patterns in data. In the context of DNA sequences, RNNs, and CNNs can be used to predict the performance of a specific sequence based on its structure and characteristics [62]. For example, RNNs can be trained on a set of DNA sequences and their corresponding performance levels and then used to predict the performance of new, unseen sequences. Similarly, CNNs can be trained to identify spatial patterns in DNA sequences that are associated with high or low performance. By combining the strengths of both RNNs and CNNs, researchers can develop more accurate and effective models for predicting the performance of DNA sequences. This can have important implications for fields such as genetic engineering and biotechnology, where the ability to accurately predict the performance of DNA sequences is crucial for developing new treatments and therapies [63].
2.3.5 Biomedical image classification applying ResNet and transfer learning
In the field of medical image analysis, one of the challenges is to accurately classify biomedical images such as X-rays, MRI scans, and CT scans, which require the expertise of trained radiologists. With the advent of DL, CNN has been widely used to classify medical images automatically. One of the most successful CNN architectures is Residual Network (ResNet), which is known for its ability to train deep networks with many layers. Transfer learning is a technique that uses pre-trained models on large datasets to solve similar tasks on smaller datasets. In biomedical image classification, transfer learning can be used to leverage pre-trained ResNet models on large datasets such as ImageNet to improve the performance of medical image classification. A pre-trained ResNet model is used as a feature extractor to apply transfer learning with ResNet in biomedical image classification [64]. The last few layers of the ResNet model, responsible for the final classification, are replaced with new layers trained on the biomedical dataset. The new layers learn the specific features of the biomedical images and improve classification accuracy. This approach has been used in various biomedical image classification tasks, such as breast cancer detection, brain tumor segmentation, and lung nodule detection, and has shown promising results in improving the accuracy of classification compared to traditional ML algorithms [65].
2.3.6 Graph embedding using GCN for protein interaction prediction
Proteins interact with each other in complex ways to perform vital biological functions. The prediction of novel protein interactions is important for understanding cellular processes and developing new drugs. Graph Convolutional Networks (GCNs) are a type of DL algorithm that can learn to represent and analyze complex network data, such as protein–protein interaction networks. In this context, GCNs can be used to perform graph embedding, which is the process of transforming the nodes and edges of a graph into a low-dimensional vector space while preserving the structural information of the graph. By using GCNs to learn the embeddings of proteins and their interactions in the network, researchers can capture the underlying patterns and relationships that are difficult to detect using traditional methods [66]. The GCN-based approach for predicting protein interactions involves training a model on a graph representation of known interactions, where the nodes represent proteins and the edges represent their interactions. The model then learns to predict whether a new interaction exists between two proteins based on their embedding vectors. One of the advantages of this approach is that it can incorporate additional features, such as protein sequence and structure information, to improve the accuracy of the predictions. Transfer learning techniques can also be used to improve the performance of the model by leveraging pre-trained embeddings from related tasks. Overall, the use of GCNs for graph embedding and predicting protein interactions has shown promising results and has the potential to contribute to the development of new drugs and therapies [67].
2.3.7 GAN image super-resolution in biology
GAN image super-resolution in biology is a DL technique used to enhance the resolution of biological images such as microscopy or medical images. GANs are composed of two neural networks: a generator and a discriminator network. The generator network generates a high-resolution image from a low-resolution input image, while the discriminator network determines whether the generated image is real or not. In GAN image super-resolution, the generator network takes a low-resolution image as input and generates a high-resolution image that is similar to the original high-resolution image. The discriminator network evaluates the similarity between the generated and original images. The generator network is trained to generate images that fool the discriminator network into thinking they are real high-resolution images. This training continues until the generator network produces high-quality images indistinguishable from real high-resolution images. GAN image super-resolution in biology has many applications, such as enhancing the resolution of microscopy images to improve the accuracy of image analysis and improving the resolution of medical images to aid in diagnosis and treatment [68].
2.3.8 Variational autoencoder high-dimensional biological generative and data embedding
VAE stands for Variational Autoencoder, which is a type of deep generative model used in ML. It is commonly used in high-dimensional data analysis and representation learning. In the context of bioinformatics and medical informatics, VAE can be used for biological data embedding and generative modeling. In VAE, the input data are first encoded into a lower-dimensional space, called the latent space, which captures the essential features of the input data. Then, a generative model is trained to map the latent space back to the original data space, allowing for the generation of new data samples. VAE is a probabilistic model, which means it can also be used for data imputation and anomaly detection. VAE has several advantages over other generative models, such as its ability to handle missing data and its ability to learn a smooth and continuous latent space representation of the input data. It is particularly useful in high-dimensional biological data analysis, where the number of features is very large, and the data are often noisy and incomplete. In summary, VAE is a powerful tool in DL and ML for high-dimensional biological data embedding and generative modeling. It has a wide range of applications in bioinformatics and medical informatics, such as data imputation, anomaly detection, and drug discovery [69]. In the next section, we delve deep into some related survey papers investigating this area.
3 Relevant reviews
We discussed the background and related ideas in-depth in the preceding section. In this section, we provide some significant relevant works in this area. In this regard, Li, Huang [19] proposed a comprehensive review of the recent developments in DL techniques for bioinformatics. They discussed the importance of big data in bioinformatics and the potential of DL techniques to analyze and make predictions based on such data. The paper provided an overview of the applications of DL in various fields of bioinformatics, including gene expression analysis, protein structure prediction, drug discovery, and disease diagnosis.
Moreover, Rezende, Xavier [70] presented a comparative study of hierarchical ML algorithms for classifying biological databases. They evaluated the performance of four different algorithms, namely random forest, Naïve Bayes, decision tree, and k-nearest neighbor, in terms of their accuracy, precision, recall, and F1 score. They also compared the performance of these algorithms with a baseline non-hierarchical ML algorithm. Also, to fulfill the lack of guidelines for hierarchical data classification, Yi, You [71] provided an overview of the recent advancements in graph representation learning for bioinformatics. They discussed the growing significance of graph-based data in bioinformatics and how graph representation learning can be used to extract valuable features and knowledge from such data. They reviewed the various graph representation learning models and their advantages and limitations in bioinformatics applications.
Besides, Sharma [72] provided an in-depth review of the applications of cluster analysis in bioinformatics. They discussed the increasing importance of cluster analysis in various fields of bioinformatics, including gene expression analysis, protein structure prediction, and disease diagnosis. They provided a comprehensive overview of the different types of clustering algorithms, including hierarchical, partitioning, density-based, and model-based clustering, and their advantages and limitations in bioinformatics applications. Also, Serra, Galdi [73] provided an overview of the recent developments in ML techniques for bioinformatics and neuroimaging. They discussed the increasing importance of big data in these fields and how ML techniques can be used to analyze and make predictions based on such data. Their paper provided a comprehensive review of the applications of ML in various fields, including gene expression analysis, protein structure prediction, drug discovery, and brain imaging analysis. However.
For this reason, there is a need for a new review article on DL in bio- and medical informatics as prior studies have offered a wide overview of DL applications in other domains but have not completely explored the potential of DL in tackling the issues faced in the sector. Recent developments in DL algorithms have also opened up new possibilities for enhancing the precision and effectiveness of medical diagnosis and therapy. We intend to emphasize the areas that require more investigation and offer direction for future work in the field by offering a thorough analysis of the most recent advancements in DL and its applications in bioinformatics, molecular biology, healthcare, and genomics. Additionally, we promote the use of DL in the medical area, enhancing patient outcomes and advancing precision medicine. Table 1 contains a summary of relevant works.
4 Methodology of research
To clearly understand ML application in bio- and medical informatics, an SLR mechanism is used in this part which is a significant survey and study of all research on a definite area. This evaluation is applied to fulfill an in-detailed examination of the DL mechanism application and explore the validity of the study selection strategy. The further subsections elaborate on the investigation process, containing research questions and criteria of paper choice.
4.1 Formalization of question
The main goals of this research are to review, classify, detect, and analyze several pertinent papers explored in ML applications in bio- and medical informatics. To gain the targets mentioned, the facets and characteristics of the mechanisms can be studied properly by applying an SLR. An even more purpose of SLR is to identify the major topics and difficulties this section addresses. The following topics are short Research Questions (RQs) that have been developed:
-
RQ 1: How may DL approaches in bio- and medical informatics be classified in medical healthcare? What are some of their examples?
This question is answered in Sect. 5.
-
RQ 2: What are the most significant cutting-edge works? What are their benefits and drawbacks? What features do they have?
Sections 5 .1 through 5.7 provide answers to this question.
-
RQ 3: What are the most widely utilized applications, techniques, criteria, and other factors in bio- and medical informatics?
This is addressed in part 6
-
RQ 4: What are the key potential solutions and unanswered issues in this field?
Part 5 will review the answers to this topic, while Part 7 will review the remaining concerns.
4.2 The procedure of paper exploration
This investigation comprises a four-stage process for exploring and selecting papers, as demonstrated in Fig. 1. Table 2 displays the terms and keywords used to explore the articles in the first phase, which were discovered through a search of traditional electronic databases such as Google Scholar, Scopus, ACM, Springer Link, Elsevier, Emerald insight, Taylor and Francis, IEEE Explore, MDPI, Wiley, and DOAJ, as well as papers, chapters, journals, books, conference papers, notes, special issues, and technical studies. The first phase yielded 790 articles, with Fig. 2 showing the distribution of articles by the publisher. In Phase 2, two phases were used to specify the total number of articles to investigate. Firstly, the involved criteria in Fig. 3 were utilized, which resulted in 467 articles remaining. Figure 4 shows the dispersion of articles by the publisher, while Fig. 5 depicts the first phase.
The survey papers are exploited in phase 3, out of 211 remaining papers in the former phase. Most of the used articles were published by Elsevier (38.5% percent). At this stage, 46 papers were remaining. The abstract and conclusion of the papers were studied in the fourth phase. Hence, 25 articles that satisfied the requirement for the precise criteria were chosen to be used and examined. In the third step, Fig. 6 displays the dispersion of the selected articles by their publishers in the second phase. Figure 7 depicts the journals that publish papers in the third phase. Table 3 indicates the specifications of the selected papers.
5 DL approaches in the field of bio- and medical informatics
This part discusses the ML mechanisms for detecting and assessing bio- and medical informatics and relevant situations. 25 articles were investigated in this part, all of which met the demand for selection criteria. To begin with, the methods were divided into 5 major classes: CNNs, RNNs, GANs, MLPs, and hybrid methods, synthesizing mechanisms. Figure 8 displays the proposed taxonomy of ML/DL methods for bio- and medical informatics.
5.1 CNN approaches for bio- and medical informatics
CNN is a fundamental DL approach that has been employed in practically all areas of medicine and is one of the useful methods for researchers. The technique is prevalently utilized for identifying MRI and CT scan images, and relevant backgrounds, as debated in the second part. In this regard, Liu, Xu [74] presented an intelligent dental health-IoT system based on smart hardware, DL, and a mobile terminal to assess its potential in in-home dental healthcare. Moreover, sophisticated dental equipment is being developed and upgraded to operate the image attainment of teeth. Based on a dataset of 12,600 clinical images collected by the presented device from 10 private dental clinics, an automatic detection model trained by MASK R-CNN was improved for the identification and classification of 7 different dental diseases, including deteriorated teeth, periodontal disease, fluorosis, and dental plaque, with detection precision of up to 90% and high specificity and sensitivity. Following a one-month assessment in ten clinics compared to the previous month, when the platform is not used, the average detection time for each patient lowers by 37.5%, demonstrating an 18.4% improvement in the treated patients.
Also, Nematzadeh, Kiani [75] presented a metaheuristic-based approach for optimizing the hyperparameters of ML algorithms and DNNs in bioinformatics applications. They discussed the challenges of selecting appropriate hyperparameters and the limitations of existing methods. They proposed a metaheuristic-based approach that involves the use of different optimization algorithms to search the hyperparameter space and identify the optimal combination of hyperparameters that leads to the best performance.
By the same token, Chen, Wang [25] evaluated nhKcr on a benchmark dataset and compared its performance with four state-of-the-art crotonylation site predictors. They tested the performance of nhKcr on a dataset commonly used to evaluate the accuracy of crotonylation site prediction tools. They also compared the performance of nhKcr to four other prediction tools that are currently considered to be the most accurate. The results showed that nhKcr outperformed the other predictors in terms of both prediction accuracy and execution time. Their results demonstrated the potential of DL-based methods for predicting post-translational modifications on nonhistone proteins.
Also, Kumar and Sharma [76] illustrated the efficiency and robustness of the COVID-19 patient’s technique of non-contact examination, which can aid in cost-efficiency and early screening and diagnosing of COVID cases. They provided images of Grad’s chest radiographs as well as the regions of interest for proven COVID-19-positive patients, bacterial pneumonia, and healthy cases. They also discussed the challenges faced in applying DL in bioinformatics, such as the need for large datasets, interpretability, and data quality.
Moreover, Jia, Chen [77] presented a DL and bioinformatics-based approach for the identification of breast cancer cases. The authors used a dataset consisting of 212 breast cancer patients and 212 healthy controls. The transcriptome data of these samples were analyzed using bioinformatics tools to identify Differentially Expressed Genes (DEGs). The DEGs were then used as input for a DL algorithm, which was trained to classify the samples as cancerous or non-cancerous. The authors reported high accuracy and specificity in the classification of the samples using this approach. Table 4 indicates the techniques, properties, and characteristics of CNN-informatics methods.
5.2 GAN approaches for bio- and medical informatics
It is worth noting that the GAN is the most widely used image classification and identification algorithm. It is now a well-known approach for usage in medicine and healthcare, and it is one of the most appealing strategies for investigators. In this section, we went through several various approaches in this area. To name but a few, Pastorino and Biswas [78] described a study that aims to address data privacy concerns while classifying chest X-ray images for COVID-19 detection. The authors developed a semi-supervised GAN that uses a small set of labeled data and a large set of unlabeled data to learn the features of chest X-ray images. To ensure data privacy, the authors introduced a data-blinding technique to remove personal information from the images, which may lead to data adequacy bias. They evaluated their method on a publicly available dataset and found that it achieved comparable performance to state-of-the-art methods while preserving data privacy.
Also, Auwul, Rahman [79] discussed using bioinformatics and ML approaches to identify potential drug targets and pathways for COVID-19. Using bioinformatics tools, they analyzed genomic and proteomic data of SARS-CoV-2 and its interaction with human proteins. They used ML algorithms to predict potential drug targets and pathways that could be used to treat COVID-19. The results showed several potential drug targets and pathways, including the renin–angiotensin and interferon signaling pathways.
Also, Lan, You [80] discussed using GAN in biomedical informatics. They provided a GAN framework and its applications in various areas such as image generation, data augmentation, disease diagnosis, drug discovery, and medical image analysis. Their method could detect Alzheimer’s disease (AD) on T1 scans at a very early phase with a zone under the curve of 0.727 and AD at a late phase with an area under the curve (AUC) of 0.894 and diagnose brain metastases on T1c scans with AUC 0.921.
Besides, Han, Rundo [81] proposed an unsupervised medical anomaly detection model called MADGAN, which is based on the GAN architecture. MADGAN can reconstruct multiple adjacent brain MRI slices from a single slice and generate realistic brain images. The proposed method can detect anomalous brain regions by comparing the reconstructed slices with the original ones. The authors evaluated the performance of MADGAN on two public brain MRI datasets and compared it with several state-of-the-art methods. The results showed that MADGAN outperformed other methods in terms of anomaly detection accuracy and computational efficiency, demonstrating the potential of MADGAN in medical anomaly detection tasks.
In addition, Balogh, Benczik [82] proposed a GAN model called TopoGAN for efficient link prediction in the protein–protein interaction (PPI) network. The model utilized the topological information of nodes and their neighbors to generate new nodes, which were then used to predict missing links in the network. The proposed model was evaluated on five benchmark PPI datasets and achieved superior performance compared to state-of-the-art methods. TopoGAN also showed its capability in identifying new PPIs between proteins, which were later validated by experiments. The results demonstrated the effectiveness of the proposed approach in predicting PPIs and can be useful for drug discovery and disease diagnosis. Table 5 indicates the techniques, properties, and characteristics of GAN-informatics methods.
5.3 RNN approaches for bio- and medical informatics
The RNN technique, which has been very practical in medicine and healthcare, is one of the most popular techniques for investigators. As mentioned before, it is the most conventionally applied technique for forecasting and prediction whereby we dwell deep into five approaches of this technique in this part. In this regard, Giansanti, Castelli [83] compared the performance of two different ML approaches—DL and classical ML—for the task of miRNA-target prediction. The authors used two different datasets, one containing experimentally validated miRNA-target interactions and another containing predicted interactions from multiple algorithms. They then trained and evaluated several models on these datasets, including a DNN, a random forest, a support vector machine, and a logistic regression model. Their results showed that DL models outperformed classical ML models in terms of both accuracy and area under the curve (AUC) metrics.
Moreover, Lyu, Chen [84] proposed an RNN framework according to word embedding and character representation. In the statements that are proper for the work and could be constructed by bidirectional difference and long short-term memory (LSTM) units, the authors used a conditional random field (CRF) layer and contextual data from both long-domain and directions dependencies. Their neural network model could be used for BNER without the need for human feature engineering. Based on their experimental findings, the domain-specific pre-trained word embedding and character-level representation may be used to create the function of the LSTM-RNN approaches.
Well, ElAbd, Bromberg [85] explained that amino acid sequences can be represented using a one-hot encoding scheme, where each amino acid is represented by a vector of binary values, with a “1” in the position corresponding to the amino acid and “0”s elsewhere. They demonstrated that this encoding scheme outperforms one-hot encoding in terms of accuracy and generalizability in various tasks, including protein classification and protein–ligand binding prediction. Their paper also provided a detailed description of the development and testing of the proposed encoding scheme, including evaluating different DL algorithms and hyperparameters. They concluded that their encoding scheme has the potential to improve the accuracy and efficiency of DL applications in the field of bioinformatics.
Furthermore, Liu and Gong [86] proposed an enhanced LSTM model that incorporates residual connections and attention mechanisms to improve the accuracy of the predictions. They demonstrated the effectiveness of their model using a dataset of protein–protein interaction residue pairs and compared their results to other commonly used methods. They concluded that their model outperformed other methods in terms of accuracy and computational efficiency, making it a promising tool for future research in protein–protein interactions. Afterward, the authors utilized it to anticipate protein–protein interaction interference with residue pairs and gained an appropriate accuracy of nearly 72%.
Additionally, Wang, Zeng [87] developed a CNN-based model, MusiteDeep, which takes amino acid sequences as input and predicts the phosphorylation sites with high accuracy. They tested their model on both general phosphorylation sites and kinase-specific phosphorylation sites and compared it to other commonly used methods. They found that their model outperformed other methods in terms of prediction accuracy and latency. Their paper provided a valuable tool for predicting phosphorylation sites in proteins and can contribute to the development of new therapies and treatments for diseases related to protein phosphorylation. As compared to other popular methods on the benchmark data, it achieved more than 50% relative development in the zone under the precision-recall curve in general phosphorylation site forecasting and obtains competitive results in kinase-specific anticipating. Table 6 indicates the techniques, properties, and characteristics of RNN-informatics methods.
5.4 MLP approaches for bio- and medical informatics
MLP has been specified as a broadly utilized and efficient ML mechanism recently used to detect classification based on high-dimensional genomic data. In this regard, Zhao, Shao [88] evaluated the performance of various models, including decision trees, logistic regression, and neural networks, in terms of both prediction accuracy and interpretability. They found that explainable ML models, such as decision trees and logistic regression, provided better interpretability than more complex models like neural networks while maintaining similar prediction accuracy. They also proposed an optimization method to improve the performance of explainable ML models.
By the same token, an IoT-based health monitoring model was suggested by Souri, Ghafour [89] to control critical signs and identify biological and behavioral alternations of learners by intelligent student care technologies. They proposed a system that collects data from wearable devices, such as smartwatches, and applied ML algorithms to analyze the data and diagnose the students’ health condition. They used a dataset collected from real-world experiments to evaluate the performance of their system and compared it with other commonly used methods. The results showed that their system achieved high accuracy in diagnosing health conditions, and outperformed other methods in terms of efficiency and cost-effectiveness.
Moreover, D’Orazio, Murdocca [90] introduced an MLP platform for the phenomics study of cancer cells reply of treatment, using and synthesizing the possibility of time-lapse microscopy for cell manner data achieving and robust DL software models for the hidden phenotypes extraction. They used a combination of DL and time-lapse-microscopy to monitor the growth and response of cancer cells to drugs over time. They collected a large dataset of time-lapse microscopy images and used DL models to identify and track the cells, extract features, and predict drug response. They evaluated the performance of their MLP approach and compared it with other commonly used methods. The results showed that the MLP approach achieved high accuracy in predicting drug response and outperformed other methods in terms of sensitivity and specificity.
Besides, Karim, Beyan [91] selected cancer genes to classify cancer accurately owing to emitted genes from microarray having many noises. They strived to find many characteristics and classifiers utilizing three benchmark datasets to systematically assess the functions of the characteristic selection mechanisms and ML classifiers. Also, they synthesized the classifiers to develop the function of the classification. Tested results demonstrated that the ensemble with some basis classifiers generates the best recognition rate on the benchmark dataset.
Also, AYDIN [92] trained and compared six ML architectures, called RF, Naïve Bayes (NB), LR, K-nearest neighbor (KNN), MLP, and SVM, for the detection of T4SEs utilizing 10 types of chosen characteristics and fivefold cross-validation. According to their results: (1) involved various but supplementary characteristics generally increase the predictive function of T4SEs, (2) the majority voting technique propelled to a more consistent and precise classification function while forecasting an ensemble learning architecture with customized exclusive single features. (3) Ensemble methods, gained by incorporating exclusive single-characteristic methods, display a particularly developed predictive function. Table 7 indicates the techniques, properties, and characteristics of MLP-informatics methods.
5.5 Hybrid approaches for bio- and medical informatics
Hybrid methods are one of the most complicated methods used in the medical and bioinformatics area. These techniques contain two or more methods for coping with hardships. In this study, we defined the evaluated methods which were created by applying methodologies. It is a conventionally utilized method in a diverse domain relevant to this subject. Considering this matter, Mohamed Shakeel, Baskar [93] stated that existing approaches for maintaining security and privacy in healthcare systems often fall short due to factors such as complexity, human errors, and the constant evolution of new threats. The proposed DQN approach aimed to address these issues by providing a more automated and adaptive security system. Their approach involved using DQNs to learn optimal policies for decision-making in various healthcare scenarios. The authors presented experimental results showing the effectiveness of the proposed approach in identifying and mitigating threats in healthcare systems. Their paper presented an interesting application of DL and reinforcement learning techniques for addressing security and privacy concerns in healthcare systems.
Besides, Huang, Shea [94] discussed a federated ML approach for predicting hospital stay time and mortality using distributed EMR from multiple hospitals. Their proposed patient clustering method groups similar patients based on their medical histories and diagnoses and then trains a local ML model for each cluster. These models are aggregated to create a global model that can make predictions for patients across all hospitals. Their approach was tested on a large dataset from multiple hospitals and compared to other ML models. The results showed that patient clustering improves the efficiency and accuracy of the federated ML approach, leading to better predictions for hospital stay time and mortality.
As well, Wang, Jiang [95] proposed a method for efficiently verifying the results of Genome-Wide Association Studies (GWAS) that are outsourced to a third-party cloud server for computation. The proposed method uses Zero-Knowledge Proofs (ZKP) to ensure the integrity and confidentiality of the outsourced computation. Specifically, they introduced a new ZKP scheme called the “range-and-sum ZKP,” which allows efficient verification of the correctness of the computation without revealing any sensitive information. They also provided a theoretical analysis of the proposed scheme and demonstrated its effectiveness through experiments using real GWAS datasets. Their method could be useful for ensuring the reliability and security of outsourced GWAS computations, which are becoming increasingly common in biomedical research.
Moreover, Cui, Zhu [96] proposed a federated learning framework called Federated electronic medical record with Anonymous Random Hybridization (FeARH) for privacy-preserving healthcare data analysis. Their framework is designed to protect the sensitive healthcare data of patients while allowing the model to learn from distributed EMR across multiple institutions. FeARH integrated three privacy-preserving techniques: differential privacy, random hybridization, and federated learning. The differential privacy mechanism preserved the privacy of individual records by adding random noise to the data. Random hybridization allows data from different sources to be combined randomly without exposing the original data. The results showed that FeARH achieves high prediction accuracy while preserving the privacy of the patient’s data.
In addition, Shahid, Nasajpour [30] analyzed the recent advances in ML research aimed at combating COVID-19. The authors highlighted the critical role that ML techniques have played in addressing various challenges posed by the pandemic, including virus detection, spread prevention, and medical assistance. Their paper discussed different approaches that have been used to address these challenges, such as developing predictive models for disease spread and severity, identifying risk factors associated with the disease, and developing methods for analyzing medical images and data. Table 8 indicates the techniques, properties, and characteristics of MLP-informatics methods.
After evaluating various studies conducted in DL methods for bio- and medical informatics, in the next section, we will analyze the results of our investigation and assess proposed studies to draw a well-organized evaluation.
6 Results and comparisons
In the preceding section, we examined in-depth DL/ML techniques for bio- and medical informatics. In this part, we go over the findings in great detail and look at the approaches from several perspectives. This investigation identifies various innovative applications that exhibit this technique. Augmenting knowledge in domains such as protein structure prediction, image classification, and data retrieval poses a challenge. We posit that reducing information to input tensors and tasks to training variations confers a well-structured foundation that can extend numerous indicators of progress in ML through frameworks. A key objective of this study was to motivate readers to exercise control over how data are inputted into ML models and to enhance training problems. In terms of learning, we primarily concentrated on the aforementioned categories. Additionally, we urge scholars to delve deeper into these subjects. Our survey evaluation revealed that most medical and bioinformatics investigations focused on a select blend of learning tasks or the improvement of annotation protocols and new datasets. ML has garnered significant popularity and acceptance, particularly for its implementation with CNN methods, which have demonstrated excellent results. However, there exist certain limitations to achieving the same level of efficacy in medical and bioinformatics applications. In general, research in this area is still ongoing. One of the most salient issues is the scarcity of large datasets containing high-quality patterns for training purposes. In such cases, data integration may be viable for amalgamating information from multiple sources. It is noteworthy that as the scale of data increases, so does the necessity for larger datasets to ensure that ML produces dependable results.
6.1 Analysis of results
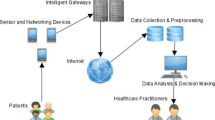
In the field of DL applications in IoT-based bio- and medical informatics, the analysis of various research papers reveals interesting findings, as depicted in Figs. 9, 10, and 11. Figure 9 presents a geo-chart showcasing the countries involved in the studied research papers. Notably, China emerges as the most prominent contributor in this field. This suggests that China has been actively engaged in research and development activities related to Deep Learning applications in IoT-based bio- and medical informatics. This could be attributed to factors such as China’s emphasis on technological advancements, significant investments in research and development, and collaborations between academic institutions and industries. Moving on to Fig. 10 illustrates the distribution of simulation environments used in DL-based approaches within the medical and bioinformatics domains. Python, a widely adopted programming language, is prominently featured, along with its popular library TensorFlow, which is widely used for implementing DL models. The popularity of Python can be attributed to its versatility, simplicity, and extensive libraries and frameworks available for DL and scientific computing. TensorFlow’s popularity stems from its powerful tools and resources for efficient DL model implementation. On the other hand, the relatively lower adoption of N2 as shown in the figure suggests its limited usage, possibly due to factors such as resource availability or compatibility issues. Finally, Fig. 11 showcases the frequency of methods applied to address medical and bioinformatics issues using DL. CNN, RNN, and GANs receive the most attention in this field. CNNs excel in image-based tasks, RNNs are suitable for processing sequential and temporal data, and GANs are promising to generate synthetic medical data and detect anomalies. The popularity of these methods indicates their effectiveness in tackling various challenges in medical and bioinformatics domains. These findings collectively highlight the contributions of different countries, the prominent simulation environments utilized, and the frequently employed DL methods in the context of DL applications in IoT-based bio- and medical informatics. They provide valuable insights into the current trends, preferences, and advancements in the field, which can guide future research and development efforts. It was observed that a substantial portion of the studies provided access to the source codes, predominantly in MATLAB and Python, to facilitate reproducibility and further experimentation. MATLAB was a prevalent choice, particularly in studies emphasizing signal processing and image analysis, owing to its extensive toolboxes tailored for these domains. Conversely, Python was prominently featured in research that incorporated machine learning frameworks like TensorFlow and Keras, aligning with the broader trend in the machine learning community. Notably, several reviewed papers included code snippets and made their complete implementations available on public repositories, fostering collaborative research and knowledge dissemination in this interdisciplinary field. This availability of codes played a pivotal role in advancing the applicability and accessibility of deep learning methodologies in the context of IoT-based bio- and medical informatics.
The use of DL techniques in biomedical and health informatics is becoming increasingly prevalent. One example is developing a smart dental health-IoT platform based on intelligent hardware, DL, and a mobile terminal. The platform can monitor oral health indicators such as temperature, pH, and moisture and use DL algorithms to detect dental diseases early. In addition, other studies have explored the use of metaheuristics to optimize hyperparameters in ML algorithms and DNNs for bioinformatics applications. This approach can improve the performance of these algorithms and ultimately lead to more accurate predictions and analysis. Furthermore, there have been efforts to apply DL techniques in bioinformatics research, such as breast cancer case identification and developing new bioinformatics tools for predicting crotonylation sites on human nonhistone proteins. These studies demonstrate the potential of DL in biomedical and health informatics research and highlight the importance of further exploring and optimizing these techniques for future applications. In the field of DL applications in IoT-based bio- and medical informatics, there is a notable emphasis on the accuracy parameter in the studies conducted, as demonstrated in Table 9. This indicates that researchers prioritize achieving high accuracy levels in their models. Accuracy is a crucial evaluation metric as it measures the overall correctness of the model’s predictions, reflecting its ability to classify and identify patterns within the data correctly. However, it is important to note that precision, which represents the proportion of true-positive predictions among all positive predictions, is the parameter receiving the least attention in these publications. Precision is a critical metric, especially in medical and bioinformatics applications, as it directly relates to correctly identifying true-positive cases while minimizing false positives. Neglecting precision can lead to potential misclassifications and incorrect diagnoses, which can have significant implications in healthcare settings.
One possible explanation for the lower emphasis on precision could be the primary focus on achieving high accuracy. Researchers may prioritize overall accuracy as it provides a comprehensive evaluation of the model’s performance, considering both positives and negatives. However, precision is equally important in healthcare and bioinformatics to avoid false positives, which can lead to unnecessary treatments or interventions. Another observation is that the majority of articles in the field tend to focus on only one target criterion while neglecting others. This limitation can hinder the comprehensive evaluation of the models and their effectiveness in real-world scenarios. To gain a deeper understanding of the model’s performance, it is essential to consider multiple evaluation parameters such as sensitivity, specificity, recall, and precision. By considering a broader range of evaluation metrics, researchers can gain a more holistic perspective on the model’s strengths and weaknesses, enabling them to make informed decisions regarding its applicability and effectiveness in practical settings. Addressing the issue of neglecting certain evaluation criteria requires researchers to place greater emphasis on the comprehensive evaluation of their models. By incorporating multiple target criteria into their studies, researchers can provide a more thorough and robust assessment of the model’s performance, ensuring that all relevant parameters are considered. This approach will contribute to a more accurate understanding of the model’s capabilities and limitations, ultimately facilitating the development of more reliable and effective DL applications in IoT-based bio- and medical Informatics.
6.2 Exploring the Integration of ML in medical applications
As shown in Sect. 5, a study investigated the impact of data adequacy bias on a semi-supervised GAN for COVID-19 chest X-ray classification. The studied papers found that data adequacy bias can reduce classification accuracy, which must be considered when developing privacy-aware GAN models [102]. They utilized a bioinformatics and ML approach to identify potential drug targets and pathways for COVID-19 treatment. One approach integrated multiple omics data sets to construct a molecular network, which was used to identify significant gene modules. A comprehensive review of the applications of GANs in biomedical informatics showed their potential in medical image analysis and drug discovery [103]. The proposed MADGAN in Sect. 5 outperformed other anomaly detection methods and could be used for the early detection of neurological diseases. An efficient link prediction model for protein–protein interaction networks was developed using topological information in a GAN framework. The model outperformed traditional network analysis methods and could be used to identify potential drug targets for diseases associated with protein–protein interactions. These studies highlight the potential of GANs and ML in medical research, particularly in disease detection, drug discovery, and protein–protein interaction network analysis [104].
Several studies have explored the use of DL and bioinformatics together. The studied models compared DL and ML approaches for miRNA-target prediction. The DL approach was found to be more accurate and precise than the ML approach. Another studied paper proposed an LSTM for biomedical named entity recognition, which outperformed traditional ML algorithms in accuracy. Another study focused on developing an amino acid encoding method for DL applications in bioinformatics. The method improved prediction accuracy for protein sequence classification. Another study used an attention mechanism enhanced LSTM with residual architecture to predict protein–protein interaction residue pairs. The model achieved high accuracy and outperformed other state-of-the-art models [105]. Lastly, a DL framework called MusiteDeep was proposed for phosphorylation site prediction. The model performed well in kinase-specific prediction tasks and achieved high accuracy. These studies demonstrate the potential of DL in various bioinformatics applications, including miRNA-target prediction, protein sequence classification, protein–protein interaction prediction, and phosphorylation site prediction. The use of DL approaches in these tasks has shown promising results and may lead to the development of more accurate and efficient tools for bioinformatics [106].
Several studies have investigated the integration of ML with medical applications. One study evaluated the effectiveness of explainable ML models for analyzing transcriptomic data. Our study demonstrated that these models can identify significant gene signatures and provide valuable insights into disease mechanisms. As shown in Sect. 5, another study proposed an ML-based healthcare monitoring model for diagnosing the condition of students in an IoT environment. This model leveraged multiple data sources to enhance diagnostic accuracy and minimize false alarms. In a different study, an ML phenomics approach combined DL with time-lapse microscopy to monitor gene expression and drug response in colorectal adenocarcinoma cells [107]. The model achieved high accuracy in predicting drug response and could potentially be useful for drug screening. A further study introduced DL-based clustering approaches for bioinformatics that can efficiently handle large and complex datasets. The models outperformed traditional clustering algorithms and could potentially be used for various bioinformatics tasks. Another study evaluated the performance of ML and bioinformatics applications on high-performance computing systems. The study demonstrated that these applications can efficiently handle massive datasets and can benefit from parallel computing. These studies demonstrate the potential of ML in medical and bioinformatics applications, specifically in areas such as healthcare monitoring, drug screening, and transcriptomic data analysis. ML models have shown promising results and could potentially lead to the development of more accurate and efficient tools for medical and bioinformatics applications. DL techniques and the incorporation of multiple data sources in ML models can lead to more accurate predictions and enhance the performance of these models [108].
Several studies have focused on integrating medical and DL subjects to improve healthcare systems. A proposed method used learning-based deep Q-networks to maintain the security and privacy of healthcare systems. Another study aimed to improve the efficiency of federated ML by using patient clustering to predict mortality and hospital stay time using distributed electronic medical records. Another proposed an efficient verification method for outsourced genome-wide association studies. In another investigation, anonymous random hybridization was utilized with federated ML to improve the privacy of electronic medical records [109]. The COVID-19 pandemic has also increased interest in ML research for virus detection, spread prevention, and medical assistance. Despite the potential benefits of ML algorithms and models for healthcare systems, concerns about privacy and security remain, and new approaches are being developed to address these issues. To leverage the benefits of ML while protecting sensitive patient data, the use of federated ML has been explored. Overall, the presented studies highlight the potential of ML in medical applications and emphasize the need for further research to improve healthcare systems and patient outcomes [110].
6.3 Prevalent evaluation criteria
One of the well-known evaluation criteria is the F-score. The mentioned keys are applied to calculate the recall, F-score, and precision. It is worth mentioning that true positive (TP) means sick people are truly recognized as sick. False positive (FP) also means intact people are wrongly recognized as sick. Also, true negative (TN) means intact people are truly recognized as intact. Furthermore, false negative (FN) means sick people are wrongly recognized as intact. Precision demonstrates the number of true results recognized truly meanwhile recall indicates the entire entities truly recognized; these concepts are calculated as follows [111]:
6.4 Challenges of The DL applications in IoT-based bio- and medical informatics
CNNs require large amounts of labeled data to train effectively. However, in the field of bio- and medical informatics, data are often limited and difficult to collect. This can lead to overfitting, where the model becomes too specialized to the training data and cannot generalize to new data. CNNs are often considered black-boxes since they can learn complex features and relationships within the data, but it can be challenging to interpret the reasons behind the model’s decision-making process. This is especially important in the medical field where doctors and researchers need to understand the reasoning behind the model’s predictions [112]. CNNs are computationally expensive and require a significant amount of processing power. This can be a significant challenge in IoT-based bio- and medical informatics, where edge computing and resource-constrained devices are common. CNNs are sensitive to data quality and can be affected by noise, missing values, and outliers. In the medical field, data can be noisy and incomplete due to the inherent complexity of biological systems, making it challenging to build accurate models. Generalization of new data: CNNs can struggle to generalize to new data that is significantly different from the training data. New patients or diseases may present unique challenges in the medical field that the model has not been trained on. Overall, CNNs are powerful tools in bio- and medical informatics, but their effective use requires careful consideration of the challenges listed above [113].
While CNNs have shown remarkable success in various image-related tasks, including medical image analysis, they have several limitations in the context of DL applications in IoT-based bio- and medical informatics. One of the major challenges with CNNs is their limited interpretability. In medical applications, understanding the reasoning behind a prediction is important, and CNNs lack transparency in this regard [114]. It is difficult to extract meaningful insights and make informed decisions based on CNN’s predictions without understanding how the model arrived at its conclusions. Another limitation of CNNs is their tendency to overfit specific data sets. This can be particularly problematic in medical applications where data sets may be small or unbalanced. While transfer learning can somewhat mitigate this, there is a need for novel techniques to improve the generalization ability of CNNs. CNNs require a large amount of labeled data to train effectively, which can be a challenge in the medical domain, where data are often scarce and expensive to obtain. This can lead to issues such as bias and limited diversity in the data set. CNNs are primarily designed for image data, and their application to other data types, such as time-series or text-based data, is limited [115]. This can be a challenge in IoT-based bio- and medical informatics, where data may be heterogeneous and multimodal. CNNs can be susceptible to adversarial attacks, where small perturbations to the input can lead to misclassification. This can be particularly concerning in medical applications, where incorrect predictions can have serious consequences. In summary, while CNNs have shown remarkable success in medical image analysis, they have several limitations that need to be addressed to improve their efficacy in IoT-based bio- and medical informatics.
In the same context, RNNs have shown great success in various applications, including NLP and time-series analysis. However, they also have some limitations when applied to IoT-based bio- and medical informatics. RNNs have a limited memory that can make it difficult to capture long-term dependencies in sequential data. This is particularly problematic in bio- and medical informatics, where the data can be complex and interdependent [116]. RNNs are trained using backpropagation through time, which can lead to vanishing gradients. This can make it difficult for the model to learn long-term dependencies in the data. RNNs can easily overfit the training data, especially if the dataset is small. This can result in poor performance when applied to new data. Training RNNs can be time-consuming, especially if the dataset is large. This can make it difficult to deploy RNN models in real-time applications. RNNs are often referred to as “black-box” models because it can be difficult to understand how they make their predictions. This can be problematic in bio- and medical informatics, where interpretability is important for ensuring patient safety. Overall, while RNNs have shown promise in IoT-based bio- and medical informatics, their limitations need to be carefully considered when developing models for real-world applications [117].
RNNs require a large amount of labeled data to train effectively, which can be difficult to obtain in the medical domain due to privacy concerns and the limited availability of data. Limited training data can lead to overfitting, where the model performs well on the training data but fails to generalize to new data. Medical data often consists of long sequences, such as ECG signals or medical records, making it challenging to process with RNNs. Long sequences can lead to vanishing or exploding gradients, which can degrade the model’s performance. RNNs can be difficult to interpret, making understanding the model’s reasoning behind its predictions challenging. In the medical domain, interpretability is critical, and understanding the model’s decision-making process is essential to building trust in the model’s predictions [118]. Medical data can be noisy and contain variations due to differences in acquisition devices, protocols, and patient conditions. Such variations can be challenging to account for, leading to decreased performance of RNNs. On the other hand, medical data collection and annotation lack standardization, making it challenging to develop RNNs that generalize well across institutions. Medical data often suffer from class imbalance, where one class (e.g., disease-positive) has significantly fewer examples than the other class (e.g., disease-negative). This issue can lead to poor performance of RNNs and requires special attention to handle. RNNs can be computationally expensive, requiring significant computational resources to train and deploy. The limited availability of high-performance computing can hinder the development and deployment of RNNs in resource-constrained settings. The use of RNNs in healthcare raises ethical considerations, such as informed consent, privacy, and bias. Addressing these issues is essential to ensure that the use of RNNs in medical applications is ethical and fair [119]. Overall, these challenges highlight the need for careful consideration of RNNs’ application in IoT-based bio- and medical informatics and the importance of addressing the unique challenges of this domain.
As well, GAN has shown groundbreaking promise in generating realistic synthetic data, but they also have some limitations and challenges when it comes to their applications in IoT-based bio- and medical informatics. GANs require large amounts of training data to learn the underlying distribution of the data [120]. However, obtaining large amounts of annotated data can be challenging and expensive in bio- and medical informatics. This can limit the effectiveness of GANs in these applications. GANs are often used to generate images, but generating high-resolution images with fine details can be challenging. This is particularly important in medical imaging applications where fine details can be critical for accurate diagnosis. GANs are often viewed as black-boxes, meaning it is difficult to understand how they arrive at their generated output. In GAN models, the lack of interpretability can be a concern in medical applications where decisions based on generated data can also have serious consequences. GANs are well suited for generating images, but they may not be as effective for other types of data, such as time-series data or text data [121]. This can limit their applicability in certain medical informatics applications. GAN training can be unstable, with the generator and discriminator networks constantly competing with each other. This can make it difficult to achieve convergence and lead to poor quality generator output.
GANs are a popular DL technique that has shown promising results in various applications, including bio- and medical informatics. However, some challenges still need to be addressed to make GANs more effective in these contexts, particularly in cloud-based settings [122]. However, GANs rely heavily on high-quality data for training, and data quality is especially critical in bio- and medical informatics. In IoT-based bio- and medical informatics, data can be noisy, incomplete, and biased, making it challenging to train GANs accurately [123]. IoT-based bio- and medical informatics involve sensitive patient data, which must be kept private and secure. However, GANs require large amounts of data to train, which poses a risk to patient privacy and security. Therefore, robust data security measures must be in place when using GANs in this context. Researchers must find ways to make GAN models more interpretable. One of the significant challenges in bio- and medical informatics is the availability of a limited dataset. The limited data can affect the accuracy of the GAN model’s results, and in some cases, it may not be possible to train a GAN model with a limited dataset. The healthcare industry is highly regulated, and GANs must comply with regulatory requirements to be approved for use. Ensuring compliance with regulations can be challenging when working with GANs, especially when dealing with IoT-based bio- and medical informatics, where data security and privacy concerns are high. In summary, while GANs offer significant potential for IoT-based bio- and medical informatics, some challenges still need to be addressed to make them more effective and acceptable for use in this context [124]. These challenges include data quality, data privacy and security, interpretability, limited datasets, and regulatory compliance.
The MLP is a type of artificial neural network that is widely used in DL applications. While MLPs have shown promising results in various domains, including IoT-based bio- and medical informatics, they also have some limitations that need to be noted. MLPs are primarily designed to handle tabular data and are not well suited for processing sequential data. This can be a limitation in bio- and medical informatics, where sequential data such as time-series data or sequences of DNA are often used [125]. MLPs are prone to overfitting, which means they can become too specialized to the training data and fail to generalize to new data. This can be particularly problematic in bio- and medical informatics when working with small datasets, where overfitting can lead to inaccurate predictions. Considering black-box models, MLP models are not easily interpretable. This means it can be challenging to understand how an MLP arrives at its predictions. Interpretability is crucial in bio- and medical informatics, where decisions can have significant consequences. MLPs do not handle missing data well. This can be a limitation in bio- and medical informatics, where datasets can be incomplete due to various reasons, such as missing data points or unbalanced data [126]. MLPs can struggle with high-dimensional data. This can be a limitation in bio- and medical informatics, where data can be high-dimensional. While MLPs have shown outperformed results in IoT-based bio- and medical informatics, it is essential to consider their limitations and explore alternative models that can better address these challenges.
MLP is a type of feedforward neural network that is commonly used in DL applications. When it comes to IoT-based bio- and medical informatics, there are several challenges associated with using MLPs. Although, one of the biggest challenges in the field of bio- and medical informatics is the limited availability of data. Data may be scarce or difficult to obtain in many cases, making it challenging to train MLP models effectively [127]. Even when data are available, it may be of poor quality. This can be due to noise, bias, or other factors that can affect the accuracy and reliability of MLP models. Referred to as black-box, it can be difficult to understand how they arrive at their predictions. In the bio- and medical informatics field, interpretability is critical, as doctors and other medical professionals need to understand and trust the predictions made by these models. Overfitting occurs when a model becomes too complex and starts to fit the noise in the training data instead of the underlying patterns. This can be a problem in bio- and medical informatics, where models need to be able to generalize to new data. In bio- and medical informatics, many ethical considerations must be taken into account when using DL models [128]. For example, it is important to ensure that the models are not biased against certain populations or groups and that they are used responsibly and ethically. Overall, MLPs can be a powerful tool in IoT-based bio- and medical informatics, but several challenges must be addressed to use them effectively.
Besides, Medical data often suffer from class imbalance, where one class (e.g., disease-positive) has significantly fewer examples than the other class (e.g., disease-negative). This issue can lead to poor performance of DL models and requires special attention to handle [129]. Real-time data processing is crucial in some medical applications, such as monitoring critical patients. However, DL models can be computationally expensive and may not be able to process data in real time. Thus, developing efficient DL models that can operate in real time is a challenge. The lack of standardization in healthcare data collection and annotation hinders the development of DL models. Different hospitals and healthcare systems use different protocols, which makes it challenging to create models that generalize well across institutions [130]. Data sharing is crucial for improving DL models’ performance, especially in healthcare, where the amount of data is limited. However, due to privacy concerns and the lack of incentives for sharing data, sharing medical data is challenging. The use of DL algorithms in healthcare raises ethical considerations, such as informed consent, privacy, and bias. Addressing these issues is essential to ensure that the use of DL models in medical applications is ethical and fair. The development and deployment of DL models can be expensive, making it difficult to implement them in resource-limited settings. Moreover, DL models may require specialized hardware and software, further increasing their cost.
In other words, the limitations of CNN methods in this context primarily revolved around their potential inefficacy in handling small or highly specialized datasets and the challenges posed by the need for substantial computational resources for training and inference, which could be a hindrance in resource-constrained IoT and cloud environments [131]. Also, the limitations of RNN methods in this systematic literature review mainly pertain to their struggle in capturing long-range dependencies in sequential data, which is crucial in certain biomedical applications, and their computationally intensive nature, potentially posing challenges in real-time processing within resource-constrained IoT and cloud environments [132]. Also, the limitations of GAN methods in this topic primarily revolved around their complexity in training and potential instability, which may require careful tuning and substantial computational resources, potentially impeding their practical implementation in resource-constrained IoT environments, while the limitations of MLP methods in this systematic literature review primarily revolved around their relative inefficacy in handling complex, high-dimensional data and their limited capability to capture intricate relationships within biomedical datasets, potentially leading to suboptimal performance in certain applications. The limitations of hybrid methods included potential challenges in model interpretability and increased complexity in combining different deep learning techniques, which may hinder their practical implementation and deployment in healthcare IoT systems.
These additional challenges highlight the multifaceted nature of implementing DL applications in IoT-based bio- and medical informatics and emphasize the need for a collaborative and interdisciplinary approach to overcome them.
6.5 Dataset in medical and bioinformatics using DL approaches
The importance of datasets in DL applications in IoT-based bio- and medical informatics cannot be overstated. DL algorithms rely on large amounts of data to learn and make accurate predictions or classifications. In bio- and medical informatics, the availability of high-quality, comprehensive datasets is crucial for developing DL models that can accurately diagnose diseases, predict treatment outcomes, and identify potential drug targets. Furthermore, the success of DL models depends heavily on the quality and diversity of the data used to train them. A biased, incomplete, or unrepresentative dataset of the target population can lead to biased or inaccurate results [133]. Therefore, it is essential to ensure that bio- and medical informatics datasets are diverse, representative, and of high quality. Moreover, using standardized datasets is critical for facilitating comparison and reproducibility of research results across different studies. Standardized datasets enable researchers to evaluate the performance of their models against others using the same data, facilitating the development of new and improved algorithms and methodologies. In summary, high-quality, comprehensive, diverse, and standardized datasets are essential for developing and evaluating DL models in IoT-based bio- and medical informatics [134]. They provide the foundation for the accurate diagnosis and treatment of diseases and the identification of new drug targets. The application of datasets in the field of DL for IoT-based bio- and medical informatics is crucial for developing accurate and efficient models. Without standard datasets, the models cannot learn and make accurate predictions. One of the key challenges in developing DL models for bio- and medical informatics is the availability of labeled datasets. Labeled datasets are critical for supervised learning, which is the most common approach in DL. This is because DL models need large amounts of labeled data to learn complex patterns and relationships in the data. In bio- and medical informatics, these labeled datasets are often created through manual annotation or by experts in the field. Many publicly available bio- and medical informatics datasets can be used for DL applications, such as the MIMIC-III dataset for EHR, the ImageNet dataset for medical imaging, and the PhysioNet dataset for physiological signals. These datasets have been used to develop models for various applications such as disease diagnosis, drug discovery, and personalized medicine. The usage of datasets in DL applications for bio- and medical informatics also requires careful attention to data privacy and security. Patient data are highly sensitive and must be handled carefully to protect patient privacy [135]. Researchers must ensure that the datasets used for training their models comply with ethical and legal requirements and that the data are de-identified before use. Researchers must carefully select and preprocess datasets, comply with ethical and legal requirements, and handle patient data with great care to protect patient privacy.
In the realm of DL applications in IoT-based bio- and medical Informatics, the datasets employed are characterized by their substantial scale and diversity. For instance, a prominent study focused on cardiac arrhythmia detection leveraged a dataset encompassing 10,000 electrocardiogram (ECG) recordings, each spanning 10 s and sampled at a rate of 500 Hz, resulting in a total of 50,000 data points per recording. Another noteworthy dataset in neurology research consisted of 500 patients with Parkinson’s disease, yielding over 150,000 data points per patient across various sensor readings. Additionally, a comprehensive dataset for Alzheimer’s disease prediction integrated multimodal data, including structural MRI images from 1000 subjects, alongside demographic and cognitive assessments. These quantitative specifics exemplify the rich and varied nature of datasets in this field, which play a pivotal role in training and evaluating deep learning models for bio- and medical informatics applications within the IoT framework.
Recent advancements in high-throughput sequencing technology have provided the scientific community with access to vast biological datasets [135]. The increased availability of these datasets has led to the expansion of Internet web services, which enable biologists to evaluate large amounts of data online for scientific audiences. Consequently, researchers have been exploring innovative methods for interrogating, evaluating, and processing data to extract information about molecular biology, biomedicine, physiology, and electronic health records. ML has gained significant popularity in the computational biology sector due to its capacity to handle massive datasets and predict outcomes with high statistical accuracy [136]. ML algorithms are statistically based computational processes that can identify hidden models in a dataset and generate reliable statistical predictions. As such, ML has been utilized in various computational biology challenges, aiding scientists in discovering critical information about diverse aspects of biology. However, most biologists and healthcare professionals lack the requisite skills to undertake a data mining project, resulting in reluctance or avoidance of ML evaluations. In other cases, researchers may follow erroneous procedures when initiating an ML venture, resulting in flawed evaluations or a false sense of success. There are various approaches to leveraging ML in computational biology research to address these issues. Though it may seem weird, the most significant key point of ML research does not consider ML: it considers your dataset attributes and deployment. To begin, you must determine whether you have enough data to address this computational biology issue with ML [137]. Currently, in the big data age, with massive biological datasets publicly available online, this issue may look unconnected, yet it appears to be a big issue in the statistical learning community and field. Whereas collecting more information can usually be advantageous for your ML patterns, considering the least dataset size to be capable of training appropriately an ML algorithm may be tricky. Even though this is not probable, the best condition would be having a minimum of ten times as many information examples as there are data characteristics.
The second crucial aspect to consider is the structuring of the dataset. In essence, this involves converting the data attributes into a standardized range, manipulating their input features, randomly reordering the dataset instances, refining and preparing the input dataset, and incorporating innovative generated characteristics, which will ultimately decide the success or failure of an ML study in a scientific assignment [138]. Due to each dataset’s idiosyncrasies and its specific scientific domain characteristics, datasets contain information crucial to their respective fields. Additionally, datasets may contain substantial errors arising from their researchers’ lack of expertise. Moreover, human curators may not always control annotations, and some may be incorrect. Further, annotations on comparable genes from various laboratories or biological research groups may differ and contain conflicting data. Such challenges can potentially impact the efficacy of an ML mechanism application. Considering the significance and the exclusiveness of every dataset area, ML research can succeed only if an investigator vividly knows the dataset details, and it may be properly configured before executing any data mining method. Managing biological datasets correctly entails numerous steps, which are commonly grouped into a phase called data preprocessing [139].
Moreover, it is often necessary to perform feature-based normalization of numerical datasets into intervals before ML algorithm analysis to bring the entire dataset into a standardized format. Hidden semantic indexing is a data retrieval strategy that relies on this preprocessing step for predicting gene performance annotation. It is a great data preprocessing tip to start with a small-scale dataset. Biology often involves large datasets with many cases [140]. Therefore, if you have a massive dataset and your ML algorithm training is time-consuming, creating a small-scale dataset with a comparable ratio to the main dataset can significantly reduce processing time. Splitting the original large dataset allows you to assess and control your approach using a combined, limited dataset. Several datasets are available for DL applications in IoT-based bio- and medical informatics. Some of the best datasets and their applicability are presented in Table 10.
Each of these datasets has unique characteristics that make them suitable for different types of research in bio- and medical informatics. For example, MIMIC-III is well suited for research in critical care, while the NIH Chest X-ray dataset is useful for research in medical imaging. Researchers can use these datasets to develop and validate DL algorithms for disease diagnosis, prediction, and treatment. However, it is important to note that these datasets have limitations and biases that must be taken into account when using them for research.
6.6 IoT applications using DL methods in bio- and medical informatics
IoT applications employing DL methods in bio- and medical informatics constitute a transformative frontier in healthcare technology. These applications leverage the interconnectedness of devices and sensors within the IoT ecosystem to revolutionize patient care, diagnosis, and treatment. DL algorithms, renowned for their prowess in processing vast and complex data, are employed to analyze diverse biomedical data streams, including physiological measurements, medical imagery, and genomic information. This enables real-time monitoring of patient health, early detection of anomalies, and personalized treatment plans. Additionally, DL-based predictive models facilitate accurate prognostic assessments and aid in the development of precision medicine approaches [137]. Moreover, integrating DL with IoT technologies enhances data security and privacy, ensuring compliance with healthcare regulations. This synergy between DL and IoT in bio- and medical informatics holds immense potential to enhance the quality of healthcare delivery and drive innovations that could reshape the future of medical practice.
6.7 Security issues, challenges, risks, IoT, and blockchain usage
The application of DL in IoT-based bio- and medical informatics poses several security challenges and risks. In particular, processing and storing large amounts of sensitive data such as patient health information raises concerns about data privacy and security. This is especially important in the case of medical data, where the misuse or mishandling of data can lead to serious consequences for patients. One of the major challenges is ensuring the security of data transmission over networks [141]. The use of IoT devices and sensors in medical applications raises concerns about the potential interception of data by malicious actors, leading to the risk of data breaches and cyber-attacks. Moreover, integrating different IoT systems and devices creates complex interdependencies that require careful consideration to avoid security vulnerabilities [142]. Blockchain technology has been proposed as a potential solution to mitigate these challenges and risks. Blockchain technology can provide a secure and tamper-resistant mechanism for storing and sharing medical data in a decentralized manner. The use of blockchain can also ensure that data are only accessible by authorized parties, and provide a way to audit data access and usage. However, there are also challenges associated with the use of blockchain in this context. For example, there are concerns about the scalability of blockchain systems and the complexity of integrating blockchain with existing systems [143]. Moreover, the use of blockchain in medical applications raises ethical and regulatory considerations related to data ownership and consent. In summary, applying DL in IoT-based bio- and medical informatics poses significant security challenges and risks. The use of blockchain technology is a promising approach for mitigating these challenges, but it also requires careful consideration and further research to ensure its effective integration and implementation in this context. Certainly, as mentioned earlier, security is a critical concern in the context of IoT-based bio- and medical informatics applications. Since these applications involve sensitive data related to individuals’ health, any security breaches can have severe consequences [144].
One way to address security concerns is through the use of blockchain technology. Blockchain is a distributed ledger technology offering a secure and tamper-proof way to store and share data. It achieves this by using cryptographic algorithms and decentralization to ensure that the data stored on the blockchain is immutable and transparent. In the context of IoT-based bio- and medical informatics, blockchain can be used to secure the data generated by IoT devices and ensure its integrity, authenticity, and privacy. For example, blockchain can be used to create a secure and tamper-proof log of all the data generated by IoT devices, which can be accessed only by authorized parties [145]. Additionally, blockchain can implement secure and privacy-preserving data sharing mechanisms between healthcare providers and researchers. However, the use of blockchain in this context also comes with its own challenges and risks [146]. For instance, blockchain’s high computational and storage requirements may not be feasible for resource-constrained IoT devices. Additionally, blockchain’s immutability can make it difficult to correct errors or update data, which can be problematic in the context of medical data that may need to be updated or corrected over time [147]. Finally, the use of blockchain also raises concerns about data privacy and confidentiality, as it can be challenging to ensure that sensitive medical data are not shared or accessed by unauthorized parties. Therefore, while blockchain technology offers a promising solution for securing IoT-based bio- and medical informatics applications, it is important to carefully consider its application and weigh the risks and benefits before implementation.
Utilizing IoT devices in medical settings comes with its own set of security issues, risks, and challenges that need to be addressed to maintain patient confidentiality and safety. Medical data are highly confidential, and unauthorized access or alteration of such data can have severe consequences. Additionally, wireless communication channels used to transmit medical data can be intercepted by attackers, which can compromise patient privacy [148]. One of the primary security challenges in AI applications in IoT-based bio- and medical informatics is the enormous amount of data generated by IoT devices, making it difficult to secure and manage. Consequently, advanced security measures must be developed by researchers to safeguard data from unauthorized access or modification. Moreover, medical data are generated in various formats and protocols, making integration and analysis difficult. This lack of interoperability between different devices and data sources presents a significant challenge in ensuring medical data security.
Another challenge is the lack of transparency and explainability of AI algorithms employed in healthcare. Healthcare providers and patients must understand how decisions are made and why certain treatments or interventions are recommended. AI algorithms in healthcare also pose ethical and legal concerns, such as potential bias, discrimination, and accountability issues. Addressing these ethical and legal considerations is crucial to ensure AI’s fairness, transparency, and accountability in healthcare. Blockchain technology is a promising solution to these security challenges. Blockchain technology provides a decentralized, secure, and transparent way of managing and sharing data. In the IoT-based bio- and medical informatics context, blockchain can secure medical data, maintain its confidentiality, integrity, and availability, and provide a tamper-proof audit trail, thus enabling transparency and accountability in decision-making processes. Furthermore, blockchain can establish trust in medical devices and their data [149]. Its decentralized nature reduces the risk of a single point of failure, making it an ideal solution for securing medical data where trust is essential. Blockchain can also manage access to medical data securely, allowing patients to control who has access to their data and grant permission to healthcare providers to access it, thus protecting their privacy. However, blockchain technology in healthcare also presents challenges such as scalability, which requires significant computational power and storage capacity to manage large volumes of data generated by IoT devices. Additionally, the lack of blockchain interoperability standards makes it difficult to integrate different blockchain networks and medical devices.
Researchers have proposed various security mechanisms for ensuring medical data security in IoT-based bio- and medical informatics, including secure communication protocols, access control mechanisms, encryption, and secure storage. They have also developed secure data aggregation mechanisms allowing medical data aggregation from multiple sources while preserving data privacy and confidentiality [150]. To address the lack of transparency and explainability of AI algorithms, researchers have proposed the use of explainable AI algorithms and AI interpretability techniques that identify factors that contribute to the decision-making process.
6.8 Upcoming deep learning models
Several emerging DL models and techniques were gaining traction but might not have been extensively utilized in this specific context at this time. One such model is the Transformer architecture, originally designed for natural language processing tasks but showing promise in various domains beyond text analysis, including image and time-series data. Its self-attention mechanism and parallel processing capabilities might offer novel approaches for handling complex biomedical data in IoT-based systems. Additionally, few-shot learning techniques, such as meta-learning and transfer learning, were garnering interest for their potential to adapt models to new tasks with limited labeled data, which could be particularly relevant in healthcare scenarios with scarce annotated datasets. Furthermore, integrating explainable AI (XAI) techniques with DL models is an emerging trend that could provide valuable insights into the decision-making process of complex models, ensuring transparency and trustworthiness in critical medical applications. It is essential to consult the latest literature and conferences related to this field for updates on the utilization of these and other novel DL models in IoT-based bio- and medical informatics [151].
Considering the comprehensive evaluation of the studied paper in DL methods in bio- and medical informatics, there are still several open issues that we intend to discuss in the next section as well as some key research challenges and future works. Moreover, transformer architectures offer a promising solution to overcome the limitations of RNNs in DL applications within IoT-based bio- and medical informatics. Unlike RNNs, transformers do not rely on sequential processing, allowing them to capture long-range dependencies more effectively. Their self-attention mechanism enables simultaneous consideration of all input elements, making them highly adept at handling complex, high-dimensional data prevalent in biomedical applications [152]. This characteristic facilitates robust feature extraction, crucial for image recognition and time-series analysis tasks. Additionally, transformers demonstrate superior parallelizability, leading to faster training times and more efficient utilization of computational resources. This attribute is particularly advantageous in resource-constrained IoT environments where real-time processing is paramount. Furthermore, transformers have demonstrated impressive performance in various natural language processing tasks, suggesting their adaptability to various data modalities. As such, incorporating transformer architectures into DL applications in IoT-based bio- and medical informatics holds great promise for advancing the state-of-the-art in this field.
7 Open issues and key challenges
In the previous section, we thoroughly examined the results. In this part, we look into open concerns and important challenges in-depth. The bioinformatics sector is a reliable source of a vast amount of daily patient data, predominantly in the form of hard copies. However, due to technological advancements in data acquisition devices, bioinformatics organizations are now collecting data in an electronic format [153]. The utilization of bioinformatics data analytics has the potential to bring about significant changes in the healthcare industry, enabling improvements in the diagnostic process and overall quality of care. Despite the considerable success of DL in various fields, such as protein structure prediction and genome editing, its application in computational biology has been met with significant challenges. DL methods often encounter problems related to a lack of annotated information, a lack of ground truth for non-simulated datasets, and significant discrepancies between training data diffusion and real-world test data diffusion, which can hinder result interpretation and benchmarking. Moreover, the use of DL methods raises ethical and moral challenges related to biases in architectures and datasets [154]. The increase in DL methods and data has made training efficiency a primary bottleneck for further advancements in the field. DL models are often regarded as inscrutable due to their lack of interpretability, posing significant challenges in medical applications where clinicians need to comprehend how the models arrived at their diagnoses or treatment recommendations. Ongoing research is focused on developing more interpretable DL models. Moreover, IoT-based bio- and medical informatics generate vast amounts of sensitive data, thereby presenting a significant risk of data breaches when using DL models, necessitating robust security measures to prevent unauthorized access, theft, or alteration of the data. However, the development and testing of DL models are restricted by a shortage of high-quality medical datasets. Furthermore, ethical concerns related to patient privacy, informed consent, and bias arise with the use of DL models in medical applications, necessitating the development of guidelines and regulations to guarantee ethical usage. In addition, integrating these models into clinical workflows and educating clinicians on their effective usage and result interpretation presents a significant challenge to their adoption in clinical settings. Another issue is the difficulty of DL models to generalize to new data beyond the training data, which is crucial in medical applications for generalizing to new patient populations or disease types. Addressing these open issues demands collaboration among researchers, clinicians, and policymakers [155]. If adequately addressed, DL models can revolutionize the field of IoT-based bio- and medical informatics, leading to better patient outcomes.
7.1 Key research challenges
This section focuses on key obstacles in further detail. The success of DL in different subareas of computational biology relies on various factors such as the availability and diversity of standardized supervised and unsupervised datasets, the computational nature of the problem, ML benchmarks with significant biological implications, and the software engineering infrastructure required to train DL architectures. Addressing the outstanding issues related to DL patterns necessitates the development of innovative solutions such as improving model explainability, generating actionable and comprehensible insights, mitigating the ethical issues associated with DL models, enhancing efficiency, and reducing training costs. The DL and computational biology communities are developing innovative solutions to tackle these challenges [156].
7.1.1 Explainability
Perhaps one of the most crucial limitations of DL models today, particularly for clinical and biological applications, is their lack of explainability. Unlike simpler regression models in statistics, it is challenging to demonstrate the importance and function of each network node in a DL model. The highly nonlinear decision boundaries and overparameterized nature of DNNs, which enable them to achieve high prediction accuracy, also make them difficult to characterize [157]. This lack of explainability is a significant obstacle in computational biology, where the trustworthiness of a DL model is essential for sensitive clinical decision-making applications. It is equally important to understand why a model can make accurate predictions as it is to understand how it makes those predictions in biology. For instance, in protein function and structure prediction, we must understand the policies controlling a protein’s 3D geometry and attributes. Addressing these problems is crucial for providing biological insights and making practical decisions in clinical settings.
In recent years, there have been numerous efforts in the ML community to enhance procedures for explaining “black-box” DL models. Many of these efforts have been applied to computational challenges in computer vision and biological applications. One of the approaches is activation maximization, which optimizes the model’s response by using gradient descent to offer an input that best represents a result. Normalization is done using closed-form density performances of the information or GANs that mimic information dispersion to make these inputs understandable to humans. Other techniques, such as the Taylor expansion for Fourier transform, use more direct approaches to extract insights from NN performance [158]. These explanations take the form of a heatmap that displays the importance of each input attribute. Another well-known process uses backpropagation to investigate the input features to which the output is most susceptible. These techniques have been used for cancer diagnostic prediction using DNNs, gene expression, and categorization.
7.1.2 Effective training
Perhaps one of the most crucial limitations of DL models today, particularly for clinical and biological applications, is their lack of explainability. Unlike simpler regression models in statistics, it is challenging to demonstrate the importance and function of each network node in a DL model. The highly nonlinear decision boundaries and overparameterized nature of DNN, which enable them to achieve high prediction accuracy, also make them difficult to characterize [159]. This lack of explainability is a significant obstacle in computational biology, where the trustworthiness of a DL model is essential for sensitive clinical decision-making applications. It is equally important to understand why a model can make accurate predictions as it is to understand how it makes those predictions in biology. For instance, in protein function and structure prediction, we must understand the policies controlling a protein’s 3D geometry and attributes. Addressing these problems is crucial for providing biological insights and making practical decisions in clinical settings.
Effective training is crucial in the development of DL models for IoT-based bio- and medical informatics applications. DL models require large amounts of high-quality data and sufficient computational resources to achieve optimal performance. There is often limited access to large, diverse datasets in the medical domain due to privacy and confidentiality concerns [160]. Therefore, data augmentation techniques such as image and signal processing, or the use of generative models such as GANs, can be used to increase the size and diversity of the available data. Moreover, transfer learning, a technique where pre-trained models are adapted to a specific task, can be used to train DL models in medical applications effectively. This is particularly useful in cases where the available data are limited or where there is a need for the model to be trained on multiple related tasks. Another crucial aspect of effective training is hyperparameter tuning. DL models have numerous hyperparameters that need to be set correctly to achieve optimal performance. This process can be time-consuming and requires expertise in the field. However, the use of automated hyperparameter tuning techniques such as Bayesian optimization or grid search can significantly improve the efficiency of this process [161]. In summary, effective training of DL models for IoT-based bio- and medical informatics applications requires careful consideration of data quality, computational resources, data augmentation techniques, transfer learning, and hyperparameter tuning. By using these techniques, researchers can improve the accuracy and robustness of DL models, leading to better patient outcomes.
In recent years, there have been numerous efforts in the ML community to enhance procedures for explaining “black-box” DL models. Many of these efforts have been applied to computational challenges in computer vision and biological applications. One of the approaches is activation maximization, which optimizes the model’s response by using gradient descent to offer an input that best represents a result [162]. Normalization is done using closed-form density performances of the information or GANs that mimic information dispersion to make these inputs understandable to humans. Other techniques, such as the Taylor expansion for Fourier transform, use more direct approaches to extract insights from NN performance. These explanations take the form of a heatmap that displays the importance of each input attribute. Another well-known process uses backpropagation to investigate the input features to which the output is most susceptible. These techniques have been used for diagnosing cancer using DNNs, gene expression, and categorization.
7.1.3 Data security and privacy
One of the most significant challenges facing the field of IoT-based bio- and medical informatics is ensuring the security and privacy of medical data. The data collected by IoT devices are often highly sensitive, and if it falls into the wrong hands, it could have serious consequences. Therefore, researchers must develop secure, privacy-preserving methods for collecting, transmitting, and storing medical data. This includes the use of encryption, access control, and anonymization techniques.
7.1.4 Interoperability and data integration
Another significant challenge is the lack of interoperability between different medical devices and data sources. IoT devices often generate data in different formats and using different protocols, making it difficult to integrate and analyze the data. Researchers must develop standardized data formats and protocols that enable seamless data integration and interoperability across different devices and platforms. Data integration is critical to DL applications in IoT-based bio- and medical informatics. It involves combining multiple data sources from various sensors, devices, and databases into a unified dataset that can be used to train and test DL models. In medical informatics, the data sources may include EHR, medical imaging data, clinical notes, and genomic data. Different systems may generate these data sources and may have different formats, making integration challenging. However, integrating these data sources is essential to capture the full complexity of the patient’s health status. DL models trained on integrated datasets can provide a more comprehensive and accurate understanding of patient health, enabling more personalized and effective treatments. Data integration can also lead to the development of new insights and discoveries by enabling the identification of previously unknown patterns and correlations. However, data integration also poses some challenges. One significant challenge is ensuring data quality and consistency, as data from different sources may have errors, biases, or inconsistencies. Additionally, data integration may raise privacy and security concerns, as sensitive patient data from multiple sources may be combined. To address these challenges, data integration strategies need to be carefully designed to ensure data quality and consistency, protect patient privacy and security, and enable efficient data retrieval and analysis.
7.1.5 Real-time monitoring and diagnosis
Real-time monitoring and diagnosis are crucial aspects of IoT-based bio- and medical informatics. The integration of sensors and devices with DL models enables the continuous collection and analysis of data, allowing for timely diagnosis and treatment of medical conditions. For example, wearable sensors can monitor vital signs such as heart rate, blood pressure, and oxygen saturation in real time, providing continuous data streams for DL models to analyze. These models can then identify patterns and anomalies that may indicate a potential medical issue, allowing for early intervention and treatment. Real-time monitoring and diagnosis can also improve patient outcomes by enabling personalized treatment plans. By continuously collecting and analyzing data on a patient’s condition, DL models can identify individualized treatment approaches that are tailored to a patient’s specific needs. However, there are also challenges to implementing real-time monitoring and diagnosis in IoT-based bio- and medical informatics. These include the need for secure and reliable data transmission, integrating data from multiple sources, and developing effective and interpretable DL models that can provide accurate and timely diagnoses. Real-time monitoring and diagnosis is a critical application area of DL in IoT-based bio- and medical informatics. This involves continuously collecting data from various sensors and devices, processing it in real time using DL models and providing real-time feedback to medical professionals or patients. One example of real-time monitoring and diagnosis is in wearable devices that collect data on heart rate, blood pressure, and other vital signs. DL models can analyze this data in real time and alert medical professionals if any abnormalities or anomalies are detected. This can help medical professionals make timely interventions and prevent adverse health outcomes. Another example is in medical imaging, where DL models can analyze medical images in real time and provide quick and accurate diagnoses. This can be especially useful in emergencies where quick decisions must be made based on limited information. Real-time monitoring and diagnosis have the potential to improve patient outcomes and reduce healthcare costs by enabling early interventions and preventing adverse events. However, it also presents challenges related to data privacy and security and the need for robust and reliable DL models that can operate in real time. This requires the use of high-performance computing and advanced ML techniques.
7.1.6 Predictive analytics
Predictive analytics is a type of advanced analytics that involves the use of statistical models and ML algorithms to analyze historical data and make predictions about future events. In the context of real-time monitoring and diagnosis in IoT-based bio- and medical informatics, predictive analytics can be a valuable tool for identifying potential health risks and predicting patient outcomes. DL models can identify patterns and make predictions about future health events by analyzing data from various sources, such as medical devices, electronic health records, and patient-generated data. For example, predictive analytics can be used to identify patients who are at high risk of developing a particular disease or condition, allowing clinicians to intervene early and prevent the onset of the disease. In addition to predicting future health events, predictive analytics can also be used to optimize treatment plans and improve patient outcomes. By analyzing data from previous patients with similar conditions, DL models can identify the most effective treatment options for individual patients and provide personalized treatment recommendations. Real-time monitoring and diagnosis can benefit greatly from the use of predictive analytics, as it allows clinicians to take proactive measures to prevent adverse health events and improve patient outcomes. However, it is important to note that predictive analytics is only as accurate as the data it is based on. Therefore, it is crucial to ensure that the data used for training and testing DL models is accurate, representative, and unbiased.
7.1.7 Ethical and legal considerations
The use of DL applications in IoT-based bio- and medical informatics raises ethical and legal considerations that must be addressed. One of the primary concerns is the privacy of patient data. As DL algorithms analyze large amounts of personal data, it is critical to maintain patient confidentiality. This requires strict security measures and protocols to prevent unauthorized access, data breaches, or theft. Another ethical consideration is the potential for bias in DL models. Biases can be unintentionally introduced in the training data, leading to inaccurate results or unequal access to medical care. Therefore, it is essential to develop guidelines and regulations to ensure that DL models are developed and used ethically and that patient rights are protected. Moreover, informed consent is another ethical consideration. Patients must be fully informed of the use of their data and the potential risks and benefits associated with the use of DL models in their medical care. It is essential to obtain informed consent from patients before using their data in any DL application. Lastly, there are also legal considerations related to the use of DL in medical applications. The regulations governing the use of medical data vary from country to country, and it is important to ensure compliance with these regulations. Additionally, liability issues may arise if a DL model produces incorrect diagnoses or treatment recommendations. Therefore, it is necessary to establish legal frameworks and guidelines for the development and deployment of DL applications in medical settings.
7.1.8 Human–computer interaction
Human–computer interaction (HCI) refers to the design, evaluation, and implementation of interactive computer systems that take into account the user’s needs, goals, and limitations. In the context of DL applications in IoT-based bio- and medical informatics, HCI is an essential aspect that helps ensure that the technology is usable, efficient, and effective for healthcare professionals and patients. HCI plays a crucial role in the development and deployment of DL applications in healthcare settings. It involves the design of user interfaces and interaction techniques that enable users to interact with DL models and make informed decisions based on their outputs. For example, a user interface that provides a visualization of the DL model’s output in real time could be used to facilitate the interpretation and understanding of the model’s predictions. Moreover, HCI is vital in ensuring that DL models are designed and evaluated in a way that takes into account the ethical and legal considerations of using these technologies in healthcare. This includes ensuring that the models are transparent, interpretable, and do not perpetuate bias or discrimination. Additionally, HCI can help to ensure that DL models are used in a way that respects patient privacy and confidentiality. In summary, HCI is a critical aspect of designing, developing, and deploying DL applications in IoT-based bio- and medical informatics. It helps ensure that the technology is usable, efficient, and effective for healthcare professionals and patients and is designed and used ethically and legally.
7.1.9 Scalability and generalizability
Scalability and generalizability are two important factors in the deployment of DL models in IoT-based bio- and medical informatics. Scalability refers to the ability of a system to handle increasing amounts of data, users, or processes. In the context of DL models, scalability is important because medical datasets can be quite large and complex, requiring significant computing resources to process and analyze. Therefore, it is crucial to ensure that DL models are scalable and can handle the increasing amounts of data that will be generated in the future. Generalizability refers to the ability of a model to perform well on new, unseen data. In medical applications, generalizability is critical because it is essential that models can accurately predict outcomes for new patients. DL models are often criticized for their lack of generalizability, as they may perform well on the training dataset but struggle when presented with new data. Therefore, developing DL models that are generalizable to new patient populations and disease types is important. To address these issues, researchers are exploring new DL architectures and techniques that can improve the scalability and generalizability of models. For example, transfer learning is a technique that allows models to reuse learned features from one task to another, reducing the amount of data required for training and improving generalizability. Additionally, federated learning is a technique that allows models to be trained on distributed datasets, reducing the amount of data that needs to be transferred and improving scalability. Addressing scalability and generalizability issues is crucial for successfully deploying DL models in IoT-based bio- and medical informatics [163].
7.2 Future works
In this section, we thoroughly examine future projects. As an interdisciplinary scientific field, bioinformatics has become essential in aiding the study of “omics” areas and technologies in life sciences, primarily managing and evaluating data from various “omes.” The massive influx of high-throughput biological information in recent years, due to technological advancements in “omic” areas, has highlighted the necessity and importance of bioinformatics resources for the analysis of large and complex datasets. To meet this demand, there is a significant need for a new generation of highly qualified scientists with cross-disciplinary knowledge and skills, capable of using complex systems, software, and algorithms to manage and interpret sophisticated biological data. To achieve this goal, there are various resources available, such as international bioinformatics education and training platforms, web-based courses, workshops, research conferences, and online education. However, developing countries need more creative platforms, network and web access, educational technologies, high-performance computing systems, and better funding to improve bioinformatics education. In terms of research, bioinformatics tools must be developed to handle the increasing volume of high-throughput data from metabolomics, metagenomics, span genomics, and proteomics. Efficient tools are also required for genome annotation and assembly with high accuracy, which necessitates sequencing more genomes, polyploid species, sub-genomes, single-cell genomes, and tissues to produce quality data for programming approaches and bioinformatics algorithms. In the future, ML programs will be increasingly employed for both clinical and research purposes. Although ML algorithms have shown potential in analyzing images, their effectiveness is still dependent on the availability of computing resources. Additionally, human operators need to inspect and validate the output of ML algorithms, which can be a time-consuming process.
7.2.1 Multimodal data integration
Multimodal data integration is a promising area for future work in the field of IoT-based bio- and medical informatics. With the increasing availability of diverse data modalities, there is a need for novel DL architectures that can effectively integrate and learn from multiple sources of information. Researchers can explore the development of new multimodal architectures that can handle different data types, such as imaging, genomics, and clinical data. Multimodal data integration can potentially improve the accuracy of diagnosis and treatment in medical applications. Future research can investigate the impact of multimodal data integration on different medical conditions and assess its potential benefits and limitations. Transfer learning has been widely used in DL to improve the performance of models in domains with limited data. Researchers can investigate the use of transfer learning techniques for multimodal data integration, where the knowledge learned from one modality can be transferred to another modality. As discussed earlier, interpretability is an essential aspect of DL models in medical applications. Future research can focus on developing interpretable multimodal models that can provide insights into how the model arrived at its decision by incorporating information from different modalities. The use of multimodal data in medical applications raises ethical and legal concerns related to patient privacy, data sharing, and informed consent. Future research can investigate these concerns and develop guidelines and regulations to ensure the ethical use of multimodal data in medical applications. Overall, the integration of multimodal data is a promising area for future work in the field of IoT-based bio- and medical informatics, and there is a need for novel techniques and approaches that can effectively handle diverse data modalities and improve the accuracy of diagnosis and treatment [164].
7.2.2 Federated learning
Federated learning is a promising technique that allows for distributed model training across multiple devices, without requiring data to be centrally stored. As such, it can potentially address the data privacy and security concerns prevalent in IoT-based bio- and medical informatics. Medical data are often high-dimensional and complex, making it challenging to develop federated learning algorithms that are both efficient and accurate. Future research could focus on developing federated learning algorithms that can effectively handle these complexities. Federated learning has shown promising results in certain medical applications, such as Electroencephalography (EEG) analysis and medical imaging. However, it is still unclear how well it will perform in other applications, such as genomics or clinical decision-making. Future research could investigate the performance of federated learning in different medical applications. Communication between devices in a federated learning setup must be secure to ensure patient privacy and prevent data breaches. Future research could focus on developing communication protocols that are both secure and efficient, allowing for effective federated learning across a wide range of medical applications. Medical data often come from a variety of sources and in different formats, making it challenging to integrate for use in federated learning. Future research could focus on developing techniques to address data heterogeneity, such as data normalization and data augmentation, to improve the effectiveness of federated learning. The ultimate goal of federated learning in IoT-based bio- and medical informatics is to improve patient outcomes. Future research could focus on developing frameworks for the deployment of federated learning models in clinical practice, including how to integrate them into existing clinical workflows effectively.
7.2.3 Explainable AI
Explainable AI is an important research area in the field of DL applications in IoT-based bio- and medical informatics. Researchers can work on developing new models that are inherently interpretable, such as decision trees, rule-based models, and linear models. These models can be used in conjunction with DL models to provide more transparent results. Visualization tools can help clinicians and researchers to better understand the results of DL models. Researchers can work on developing new tools for visualizing the results of DL models and explaining how they arrived at their decisions. Researchers can develop techniques to incorporate human feedback into the training process of DL models. This can help to improve the interpretability of the models and make them more useful for clinical decision-making. Researchers can work on developing standards for interpretability in DL models. This can help ensure that models are transparent and that clinicians understand how they arrived at their decisions. Researchers can evaluate the impact of interpretability on the adoption of DL models in clinical settings. This can help to identify the most effective approaches for making DL models more interpretable and useful for clinical decision-making. By addressing the issue of interpretability in DL models, researchers can help improve the trust and adoption of these models in IoT-based bio- and medical informatics.
7.2.4 Transfer learning
Transfer learning, a technique in which a model trained on one task is adapted for use on a new task, has shown great promise in medical applications. There is a growing interest in using transfer learning for medical image analysis tasks. By adapting pre-trained models on large general image datasets like ImageNet to medical imaging tasks, we can leverage the learned features and weights to improve the performance of the models on smaller medical datasets. Transfer learning has been successfully applied to NLP tasks by pre-training large language models like BERT on vast amounts of text data. There is a need for models that can understand medical language and terminologies in the medical field. Fine-tuning these pre-trained language models on medical text datasets can improve their performance on medical text classification tasks. Transfer learning has not been widely applied to time-series data in the medical field. However, with the increasing availability of wearable devices and IoT sensors that generate time-series data, transfer learning can effectively leverage pre-trained models for tasks like patient monitoring and disease prediction. In the medical field, obtaining large amounts of data from a single institution can be challenging due to privacy and security concerns. Domain adaptation techniques can be used to transfer knowledge from pre-trained models to a new dataset with a different distribution. This can be particularly useful for tasks like disease diagnosis, where the model needs to be trained on data from multiple institutions to ensure generalizability. As mentioned earlier, multimodal data integration is an essential area of research in medical informatics. Transfer learning can be used to leverage pre-trained models from different modalities to improve the overall system’s performance. For example, pre-trained models on medical images and text can be combined to create a system that can analyze both modalities simultaneously. Overall, the use of transfer learning in the DL applications in IoT-based bio- and medical informatics has significant potential to improve the performance and efficiency of the models. Future research in this area should focus on developing new transfer learning techniques that can handle the unique challenges of medical data and integrating transfer learning with other techniques like federated learning and explainable AI.
7.2.5 Personalized healthcare monitoring
Personalized healthcare monitoring is a rapidly growing area of research that seeks to provide personalized healthcare solutions to individuals. DL, coupled with the IoT and bio- and medical informatics, has the potential to revolutionize personalized healthcare monitoring. In personalized healthcare monitoring, the data comes from multiple sources such as wearable devices, medical sensors, and electronic health records. DL techniques can be used to fuse this data to comprehensively view an individual’s health status. Multimodal data fusion using DL can help improve the accuracy and reliability of personalized healthcare monitoring systems. Anomaly detection is an important aspect of personalized healthcare monitoring as it helps in identifying unusual patterns in an individual’s health status. DL techniques can be used to identify these patterns and raise alarms if necessary. This can be particularly useful in detecting chronic diseases or sudden health emergencies. Real-time monitoring of an individual’s health status can be achieved using wearable devices and IoT-enabled sensors. DL models can be deployed on these devices to continuously monitor an individual’s health status and provide real-time alerts if necessary. This can be particularly useful for elderly or high-risk patients.
DL models can be trained on large datasets of medical records to provide personalized diagnoses to individuals. These models can take into account an individual’s medical history, genetic information, and other factors to provide accurate diagnosis and treatment recommendations. Predictive analytics using DL can help in predicting an individual’s health status and potential health risks. These models can be trained on large datasets of medical records to identify patterns and predict potential health issues. This can be particularly useful in preventive healthcare. Privacy and security are major concerns in personalized healthcare monitoring. DL models can be used to ensure the privacy and security of an individual’s health data. Techniques such as federated learning can be used to train models on distributed datasets without compromising privacy. DL models are often considered “black-boxes” as they are difficult to interpret and explain. In personalized healthcare monitoring, it is important to provide explainable and interpretable models to gain the trust of patients and healthcare providers. Explainable AI techniques can be used to provide insights into the inner workings of these models. These are just some of the future works and ideas that can be explored in personalized healthcare monitoring using DL and IoT-based bio- and medical informatics. With the increasing availability of health data and the advancement of DL techniques, personalized healthcare monitoring can potentially transform how we manage our health.
7.2.6 Real-time diagnosis and treatment planning
Real-time diagnosis and treatment planning is a critical aspect of healthcare that can benefit greatly from DL and IoT-based bio- and medical informatics. DL models can be trained on large datasets of medical images and patient records to provide a real-time diagnosis. These models can be deployed on IoT-enabled devices to provide immediate feedback to healthcare providers. This can be particularly useful in emergencies where quick diagnosis is critical. DL models can be used to develop personalized treatment plans for patients. These models can take into account an individual’s medical history, genetic information, and other factors to provide tailored treatment recommendations. IoT-enabled devices can be used to monitor a patient’s response to treatment and adjust the treatment plan accordingly. Decision support systems using DL can help healthcare providers make informed decisions about diagnosis and treatment. These systems can provide recommendations based on patient data, medical guidelines, and other relevant information. Predictive analytics using DL can help in predicting a patient’s response to treatment and potential health risks. These models can be trained on large datasets of medical records to identify patterns and predict potential health issues. This can be particularly useful in preventive healthcare. DL models can be trained to analyze medical images such as X-rays, MRIs, and CT scans. These models can help healthcare providers identify abnormalities and diagnose diseases. IoT-enabled devices can be used to capture and transmit these images in real time, enabling remote diagnosis and treatment planning. Privacy and security are major concerns in real-time diagnosis and treatment planning. DL models can be used to ensure the privacy and security of patient data. Techniques such as federated learning can be used to train models on distributed datasets without compromising privacy. In real-time diagnosis and treatment planning, it is important to provide explainable and interpretable models to gain the trust of patients and healthcare providers. Explainable AI techniques can be used to provide insights into the inner workings of these models. These are just some of the future works and ideas that can be explored in real-time diagnosis and treatment planning using DL and IoT-based bio- and medical informatics. With the increasing availability of healthcare data and the advancement of DL techniques, real-time diagnosis, and treatment planning has the potential to transform the way we deliver healthcare [165].
7.2.7 Predictive maintenance of medical devices
Predictive maintenance is an important aspect of medical device management that can benefit greatly from the use of DL and IoT-based bio- and medical informatics. Predictive analytics using DL can be used to predict when medical devices are likely to fail or require maintenance. These models can be trained on large datasets of sensor data from medical devices to identify patterns and predict potential issues. DL models can be used to monitor the condition of medical devices in real time [166]. These models can analyze data from sensors such as temperature, pressure, and vibration to detect abnormalities and potential failures. Anomaly detection using DL can help identify unusual patterns in medical device data. These models can help detect issues that may not be immediately apparent to the human eye and raise alarms if necessary. Prognostic models using DL can be used to predict the remaining useful life of medical devices. These models can help healthcare providers plan for the maintenance and replacement of medical devices before they fail. Predictive maintenance scheduling using DL can help healthcare providers optimize maintenance schedules based on the predicted failure rates of medical devices. This can help reduce downtime and improve the reliability of medical devices. Fault diagnosis using DL can help healthcare providers quickly identify and diagnose issues with medical devices. These models can analyze sensor data and provide repair or replacement recommendations. Predictive maintenance models can be integrated with electronic health records to view medical device performance and patient outcomes comprehensively. This can help healthcare providers make informed medical device management and patient care decisions. These are just some of the future works and ideas that can be explored in the predictive maintenance of medical devices using DL and IoT-based bio- and medical informatics. With the increasing use of medical devices and the need for reliable and safe healthcare delivery, predictive maintenance can potentially improve healthcare systems’ efficiency and effectiveness.
7.2.8 Optimization of drug discovery
Drug discovery is a complex and time-consuming process that can benefit greatly from the use of DL and IoT-based bio- and medical informatics. DL models can be used to design new drugs based on the molecular structure of existing drugs and the desired therapeutic effect. These models can predict the interaction between drugs and target proteins, helping to identify potential drug candidates. Virtual screening using DL can help identify potential drug candidates from large databases of compounds. These models can analyze the chemical structure of compounds and predict their activity against target proteins. Toxicity prediction using DL can help identify potential safety concerns of drug candidates. These models can analyze the chemical structure of compounds and predict their toxicity based on their interaction with target proteins. DL models can be used to identify existing drugs that may be effective in treating other diseases. These models can analyze the molecular structure of drugs and predict their potential therapeutic effects against other diseases. DL models can be used to optimize clinical trial design and reduce the time and cost of drug development. These models can predict patient response to treatment and identify subgroups that are more likely to benefit from a drug. DL models can be used to develop personalized treatment plans based on an individual’s genetic information, medical history, and other factors. These models can predict the effectiveness of different drugs and help healthcare providers make informed treatment decisions. DL models can be integrated with electronic health records to provide a comprehensive view of patient health and treatment outcomes. This can help healthcare providers make informed drug treatment and patient care decisions. These are just some of the future works and ideas that can be explored in the optimization of drug discovery using DL and IoT-based bio- and medical informatics. With the increasing demand for new and effective drugs, drug discovery optimization has the potential to transform the pharmaceutical industry and improve patient outcomes.
7.2.9 Medical imaging analysis
Medical imaging analysis is a critical aspect of healthcare that can benefit greatly from the use of DL and IoT-based bio- and medical informatics. DL models can be used for image segmentation, which involves separating an image into different regions based on their characteristics. This can help identify and isolate specific structures or abnormalities in medical images. DL models can be used for image classification, which involves assigning a label to an image-based on its content. This can help identify different types of structures or abnormalities in medical images. DL models can be used for image registration, which involves aligning multiple medical images of the same patient taken at different times or from different modalities. This can help track patient condition changes over time and improve treatment planning. DL models can be used for image reconstruction, which involves creating high-quality images from low-quality or incomplete data. This can help improve the accuracy of medical imaging and reduce the need for additional imaging tests. DL models can be trained to diagnose medical conditions based on medical images automatically. This can help reduce radiologists’ workload and improve diagnosis speed and accuracy. DL models can be used for quantitative analysis of medical images, which involves measuring and analyzing different aspects of the images, such as size, shape, and texture. This can help identify subtle changes in medical images that may be difficult to detect with the human eye. DL models can be integrated with electronic health records to provide a comprehensive view of patient health and treatment outcomes. This can help healthcare providers make informed decisions about patient care. These are just some of the future works and ideas that can be explored in medical imaging analysis using DL and IoT-based bio- and medical informatics. With the increasing use of medical imaging in healthcare, medical imaging analysis has the potential to improve the accuracy and efficiency of diagnosis and treatment planning.
7.2.10 Health monitoring with wearable IoT devices and DL
Health monitoring with wearable IoT devices and DL can revolutionize healthcare by providing continuous monitoring of patient health and allowing for early detection of health problems. Continuous Vital Sign Monitoring: Wearable IoT devices can be used to continuously monitor vital signs such as heart rate, blood pressure, and respiratory rate. DL models can analyze the data from these devices to identify patterns and detect early warning signs of health problems. Wearable IoT devices can be used to monitor chronic diseases such as diabetes and hypertension. DL models can analyze the data from these devices to detect changes in disease status and provide feedback on treatment effectiveness. Wearable IoT devices can be used to monitor behavior patterns such as sleep, physical activity, and nutrition. DL models can analyze the data from these devices to identify patterns and provide feedback on lifestyle modifications. Wearable IoT devices can be used to detect falls in elderly patients and individuals with balance problems. DL models can analyze the data from these devices to detect falls and alert healthcare providers or family members. Wearable IoT devices can be used to monitor medication adherence in patients with chronic diseases. DL models can analyze the data from these devices to provide feedback on medication adherence and improve patient outcomes. DL models can be used to develop early warning systems for critical health events such as heart attacks and strokes. Wearable IoT devices can be used to monitor vital signs and detect early warning signs, allowing for prompt medical intervention. Wearable IoT devices and DL models can be integrated with electronic health records to provide a comprehensive view of patient health and treatment outcomes. This can help healthcare providers make informed decisions about patient care. These are just some of the future works and ideas that can be explored in health monitoring with wearable IoT devices and DL in IoT-based bio- and medical informatics. With the increasing use of wearable IoT devices in healthcare, health monitoring has the potential to improve patient outcomes and reduce healthcare costs.
7.2.11 Telemedicine
Telemedicine has become an increasingly popular approach to healthcare delivery, especially in remote or underserved areas. The integration of DL with IoT-based bio- and medical informatics can help improve the quality of telemedicine services and enhance patient outcomes. DL models can be trained to remotely analyze patient data such as medical images, laboratory results, and vital signs. This can help improve the accuracy and speed of diagnosis, especially in areas with limited access to healthcare professionals. DL models can be used to develop chatbots and virtual assistants that can communicate with patients and provide medical advice. This can help improve patient access to healthcare services and reduce the workload of healthcare professionals. IoT-based wearable devices can be used to remotely monitor patient health data such as heart rate, blood pressure, and respiratory rate. DL models can analyze this data in real time and alert healthcare professionals if any changes require attention. DL models can be used to analyze patient data to identify patients who are at risk of developing certain diseases. This can help healthcare professionals to provide proactive care and prevent disease progression. DL models can be used to develop personalized treatment plans based on patient data. This can help improve treatment outcomes and reduce healthcare costs by avoiding unnecessary treatments. DL models can be used to develop automated triage systems that can identify patients who require urgent care. This can help reduce wait times for patients who require immediate attention. Telemedicine services can be integrated with electronic health records to provide a comprehensive view of patient health and treatment outcomes. This can help healthcare providers make informed decisions about patient care. These are just some of the future works and ideas that can be explored in telemedicine with the integration of DL and IoT-based bio- and medical informatics. With the increasing demand for telemedicine services, the integration of these technologies has the potential to improve access to healthcare services and enhance patient outcomes.
7.2.12 Predictive analytics for healthcare
Predictive analytics has become an essential tool for healthcare providers in making informed decisions about patient care. The integration of DL with IoT-based bio- and medical informatics can help improve the accuracy and speed of predictive analytics, leading to better patient outcomes. DL models can be used to analyze patient data such as medical images, laboratory results, and vital signs to detect early warning signs of diseases. This can help healthcare providers to provide timely interventions and prevent disease progression. DL models can be used to develop predictive risk models that identify patients at high risk of developing certain diseases. This can help healthcare providers to provide proactive care and prevent disease progression. DL models can be used to develop personalized treatment plans based on patient data. This can help improve treatment outcomes and reduce healthcare costs by avoiding unnecessary treatments. Predictive analytics can be used to optimize healthcare resources such as hospital beds, staff, and equipment. DL models can be used to predict patient demand and optimize resource allocation accordingly. DL models can be used to analyze patient data to identify potential drug interactions and adverse events. This can help healthcare providers to provide safer and more effective drug therapies. DL models can be used to develop clinical decision support systems that can assist healthcare providers in making informed decisions about patient care. This can help improve patient outcomes and reduce healthcare costs by avoiding unnecessary tests and treatments. DL models can be used to analyze population health data to identify health trends and disease outbreaks. This can help healthcare providers to develop targeted interventions to prevent the spread of disease. These are just some of the future works and ideas that can be explored in predictive analytics for healthcare with the integration of DL and IoT-based bio- and medical informatics. With the increasing demand for predictive analytics in healthcare, integrating these technologies has the potential to improve patient outcomes and reduce healthcare costs.
8 Conclusion and limitation
The DL applications in IoT-based bio- and medical informatics have exhibited remarkable progress in recent years, with various studies demonstrating the effectiveness of DL in different areas such as drug discovery, disease diagnosis, and patient monitoring. Nonetheless, the field is continuously evolving, and further research is necessary to explore new techniques and methodologies that can enhance the performance and robustness of DL algorithms in the context of bio- and medical informatics. In addition, there is a need for more comprehensive evaluations of DL algorithms in real-world scenarios and for the development of robust and scalable systems that can be deployed in healthcare settings. Therefore, it is imperative to continue conducting research in this area to fully leverage the potential of DL in IoT-based bio- and medical informatics and provide better healthcare outcomes for patients. To this end, this article presents a systematic review of DL-based methods used for bio- and medical informatics issues. Initially, we discuss the advantages and disadvantages of some surveyed papers about medical and bioinformatics-related methods, before illustrating the strategy of this article. The DL-bioinformatics platforms and tools are also assessed. Based on a survey of papers according to qualitative features, most papers are assessed relying on accuracy, sensitivity, specificity, F-score, adaptability, scalability, and latency. However, certain features, such as security and convergence time, are underutilized. To evaluate and perform the proposed methods, various programming languages are used. Furthermore, we anticipate that our investigation will provide a valuable guide for further research on DL and medical usage in medical and bioinformatics issues.
Nevertheless, some constraints were encountered during our analysis, including the unavailability of non-English papers, which limited our ability to utilize numerous investigation initiatives. Additionally, some of the papers examined had significant limitations in clear explanations of the algorithms used. Finally, another limitation we faced was a shortage of availability to different papers published by significant publications.
Availability of data and materials
The paper contains all of the data.
References
Muhammad AN et al (2021) Deep learning application in smart cities: recent development, taxonomy, challenges and research prospects. Neural Comput Appl 33(7):2973–3009
Nosratabadi S et al (2020) State of the art survey of deep learning and machine learning models for smart cities and urban sustainability. In: International conference on global research and education. Springer
Shafqat S et al (2022) Standard ner tagging scheme for big data healthcare analytics built on unified medical corpora. J Artif Intell Technol 2(4):152–157
Atitallah SB et al (2020) Leveraging deep learning and iot big data analytics to support the smart cities development: review and future directions. Comput Sci Rev 38:100303
Kök I, Şimşek MU, Özdemir (2017) A deep learning model for air quality prediction in smart cities. In: 2017 IEEE international conference on big data (Big Data). 2017. IEEE
Bolhasani H, Mohseni M, Rahmani AM (2021) Deep learning applications for IoT in health care: a systematic review. Inform Med Unlocked 23:100550
Rastogi R, Chaturvedi DK, Sagar S, Tandon N, Rastogi AR (2022) Brain tumor analysis using deep learning: sensor and iot‐based approach for futuristic healthcare. In: Bioinformatics and medical applications: big data using deep learning algorithms, pp 171–190
Roopashree S et al (2022) An IoT based authentication system for therapeutic herbs measured by local descriptors using machine learning approach. Measurement 200:111484
Bharadwaj HK et al (2021) A review on the role of machine learning in enabling IoT based healthcare applications. IEEE Access 9:38859–38890
Awotunde JB et al (2021) Disease diagnosis system for IoT-based wearable body sensors with machine learning algorithm. In: Hybrid artificial intelligence and IoT in healthcare. 2021. Springer, pp 201–222
Alansari Z et al (2017) Computational intelligence tools and databases in bioinformatics. In: 2017 4th IEEE international conference on engineering technologies and applied sciences (ICETAS). 2017. IEEE
Daoud H, Williams P, Bayoumi M (2020) IoT based efficient epileptic seizure prediction system using deep learning. In: 2020 IEEE 6th world forum on internet of things (WF-IoT). 2020. IEEE
Wu Y et al (2021) Deep learning for big data analytics. Mobile Netw Appl 26(6):2315–2317
Ambika N (2022) An economical machine learning approach for anomaly detection in IoT environment. In: Bioinformatics and medical applications: big data using deep learning algorithms, 2022: pp 215–234
Srivastava M (2020) A Surrogate data-based approach for validating deep learning model used in healthcare. In: Applications of deep learning and big IoT on personalized healthcare services. 2020. IGI Global, pp 132–146
da Costa KA et al (2019) Internet of things: a survey on machine learning-based intrusion detection approaches. Comput Netw 151:147–157
Min S, Lee B, Yoon S (2017) Deep learning in bioinformatics. Brief Bioinform 18(5):851–869
Aminizadeh S et al (2023) The applications of machine learning techniques in medical data processing based on distributed computing and the internet of things. In: Computer methods and programs in biomedicine, 2023, p 107745
Li Y et al (2019) Deep learning in bioinformatics: introduction, application, and perspective in the big data era. Methods 166:4–21
Cao Y et al (2020) Ensemble deep learning in bioinformatics. Nat Mach Intell 2(9):500–508
Tang B et al (2019) Recent advances of deep learning in bioinformatics and computational biology. Front Genet 10:214
Koumakis L (2020) Deep learning models in genomics; are we there yet? Comput Struct Biotechnol J 18:1466–1473
Dhombres F, Charlet J (2019) Formal medical knowledge representation supports deep learning algorithms, bioinformatics pipelines, genomics data analysis, and big data processes. Yearb Med Inform 28(01):152–155
Peng L et al (2018) The advances and challenges of deep learning application in biological big data processing. Curr Bioinform 13(4):352–359
Chen Y-Z et al (2021) nhKcr: a new bioinformatics tool for predicting crotonylation sites on human nonhistone proteins based on deep learning. Brief Bioinform 22(6):bbab146
Chen Y et al (2016) Gene expression inference with deep learning. Bioinformatics 32(12):1832–1839
Jabbar MA (2022) An insight into applications of deep learning in bioinformatics. In: Deep learning, machine learning and IoT in biomedical and health informatics. CRC Press, pp 175–197
Khurana S et al (2018) DeepSol: a deep learning framework for sequence-based protein solubility prediction. Bioinformatics 34(15):2605–2613
Baranwal M et al (2020) A deep learning architecture for metabolic pathway prediction. Bioinformatics 36(8):2547–2553
Shahid O et al (2021) Machine learning research towards combating COVID-19: Virus detection, spread prevention, and medical assistance. J Biomed Inform 117:103751
Roy PK et al (2023) Analysis of community question-answering issues via machine learning and deep learning: state-of-the-art review. CAAI Trans Intell Technol 8(1):95–117
Samanta RK et al (2022) Scope of machine learning applications for addressing the challenges in next-generation wireless networks. CAAI Trans Intell Technol 7(3):395–418
Wang W et al (2023) Fully Bayesian analysis of the relevance vector machine classification for imbalanced data problem. CAAI Trans Intell Technol 8(1):192–205
Ashrafuzzaman M (2021) Artificial intelligence, machine learning and deep learning in ion channel bioinformatics. Membranes 11(9):672
Fiannaca A et al (2018) Deep learning models for bacteria taxonomic classification of metagenomic data. BMC Bioinform 19(7):61–76
Li F et al (2020) DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics 36(4):1057–1065
Meher J (2021) Potential applications of deep learning in bioinformatics big data analysis. In: Advanced deep learning for engineers and scientists, 2021, pp 183–193
Preuer K et al (2018) DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics 34(9):1538–1546
Xia Z et al (2019) DeeReCT-PolyA: a robust and generic deep learning method for PAS identification. Bioinformatics 35(14):2371–2379
Fang B et al (2022) Deep generative inpainting with comparative sample augmentation. J Comput Cogn Eng 1(4):174–180
Wang X et al (2020) Block switching: a stochastic approach for deep learning security. arXiv preprint arXiv:2002.07920, 2020
Kumar I, Singh SP (2022) Machine learning in bioinformatics. In: Bioinformatics. Academic Press, pp 443–456
Yu L et al (2018) Drug and nondrug classification based on deep learning with various feature selection strategies. Curr Bioinform 13(3):253–259
Jurtz VI et al (2017) An introduction to deep learning on biological sequence data: examples and solutions. Bioinformatics 33(22):3685–3690
Deng Y et al (2020) A multimodal deep learning framework for predicting drug–drug interaction events. Bioinformatics 36(15):4316–4322
Shakeel N, Shakeel S (2022) Context-free word importance scores for attacking neural networks. J Comput Cogn Eng 1(4):187–192
Oubounyt M et al (2019) DeePromoter: robust promoter predictor using deep learning. Front Genet 10:286
Leung MK et al (2014) Deep learning of the tissue-regulated splicing code. Bioinformatics 30(12):i121–i129
Dai B, Bailey-Kellogg C (2021) Protein interaction interface region prediction by geometric deep learning. Bioinformatics 37(17):2580–2588
Luo F et al (2019) DeepPhos: prediction of protein phosphorylation sites with deep learning. Bioinformatics 35(16):2766–2773
Liu X (2022) Real-world data for the drug development in the digital era. J Artif Intell Technol 2(2):42–46
Wei L et al (2018) Prediction of human protein subcellular localization using deep learning. J Parallel Distrib Comput 117:212–217
Heidari A et al (2023) A new lung cancer detection method based on the chest CT images using federated learning and blockchain systems. Artif Intell Med 141:102572
Cai Q et al (2023) Image neural style transfer: a review. Comput Electr Eng 108:108723
Ai Q et al (2021) Editorial for FGCS special issue: intelligent IoT systems for healthcare and rehabilitation. Elsevier, New York, pp 770–773
Niu L-Y, Wei Y, Liu W-B, Long JY, Xue T-H (2023) Research Progress of spiking neural network in image classification: a review. In: Applied intelligence, pp 1–25
Karnati M et al (2022) A novel multi-scale based deep convolutional neural network for detecting COVID-19 from X-rays. Appl Soft Comput 125:109109
Ravindran U, Gunavathi C (2023) A survey on gene expression data analysis using deep learning methods for cancer diagnosis. Prog Biophys Mol Biol 177:1–13
Zheng M et al (2022) A hybrid CNN for image denoising. J Artif Intell Technol 2(3):93–99
Togneri R, Prati R, Nagano H, Kamienski C (2023) Data-driven water need estimation for IoT-based smart irrigation: a survey. Expert Syst Appl 225:120194
Sheng N, Huang L, Lu Y, Wang H, Yang L, Gao L, Xie X, Fu Y, Wang Y (2023) Data resources and computational methods for lncRNA-disease association prediction. Comput Biol Med 153:106527
Sharan RV, Rahimi-Ardabili H (2023) Detecting acute respiratory diseases in the pediatric population using cough sound features and machine learning: a systematic review. Int J Med Inform 176:105093
Bhosale YH, Patnaik KS (2023) Bio-medical imaging (X-ray, CT, ultrasound, ECG), genome sequences applications of deep neural network and machine learning in diagnosis, detection, classification, and segmentation of COVID-19: a meta-analysis & systematic review. Multimed Tools Appl 82:39157–39210. https://doi.org/10.1007/s11042-023-15029-1
Azhari F, Sennersten CC, Lindley CA et al (2023) Deep learning implementations in mining applications: a compact critical review. Artif Intell Rev 56:14367–14402. https://doi.org/10.1007/s10462-023-10500-9
Nazir S, Dickson DM, Akram MU (2023) Survey of explainable artificial intelligence techniques for biomedical imaging with deep neural networks. Comput Biol Med 156:106668
Jacob TP, Pravin A, Kumar RR (2022) A secure IoT based healthcare framework using modified RSA algorithm using an artificial hummingbird based CNN. Trans Emerg Tel Tech 33(12):e4622. https://doi.org/10.1002/ett.4622
Phan HT, Nguyen NT, Hwang D (2023) Aspect-level sentiment analysis: a survey of graph convolutional network methods. Inf Fusion 91:149–172
Qiu D, Cheng Y, Wang X (2023) Medical image super-resolution reconstruction algorithms based on deep learning: a survey. Comput Methods Prog Biomed 238:107590
Sanders LM et al (2023) Biological research and self-driving labs in deep space supported by artificial intelligence. Nat Mach Intell 5(3):208–219
Rezende PM et al (2022) Evaluating hierarchical machine learning approaches to classify biological databases. Brief Bioinform 23(4):bbac216
Yi H-C et al (2022) Graph representation learning in bioinformatics: trends, methods and applications. Brief Bioinform 23(1):bbab340
Sharma S (2021) The bioinformatics: detailed review of various applications of cluster analysis. Glob J Appl Data Sci Internet Things 5:1–2021
Serra A, Galdi P, Tagliaferri R (2018) Machine learning for bioinformatics and neuroimaging. Wiley Interdiscip Rev Data Min Knowl Discov 8(5):e1248
Liu L et al (2019) A smart dental health-IoT platform based on intelligent hardware, deep learning, and mobile terminal. IEEE J Biomed Health Inform 24(3):898–906
Nematzadeh S et al (2022) Tuning hyperparameters of machine learning algorithms and deep neural networks using metaheuristics: a bioinformatics study on biomedical and biological cases. Comput Biol Chem 97:107619
Kumar H, Sharma S (2021) Contribution of deep learning in bioinformatics. Glob J Appl Data Sci Internet Things 5:1–202
Jia D et al (2021) Breast cancer case identification based on deep learning and bioinformatics analysis. Front Genet 12:628136
Pastorino J, Biswas AK (2022) Data adequacy bias impact in a data-blinded semi-supervised GAN for privacy-aware COVID-19 chest X-ray classification. In: Proceedings of the 13th ACM international conference on bioinformatics, computational biology and health informatics, 2022
Auwul MR et al (2021) Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief Bioinform 22(5):bbab120
Lan L et al (2020) Generative adversarial networks and its applications in biomedical informatics. Front Public Health 8:164
Han C et al (2021) MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform 22(2):1–20
Balogh OM et al (2022) Efficient link prediction in the protein–protein interaction network using topological information in a generative adversarial network machine learning model. BMC Bioinform 23(1):1–19
Giansanti V et al (2019) Comparing deep and machine learning approaches in bioinformatics: a miRNA-target prediction case study. In: International conference on computational science. 2019. Springer
Lyu C et al (2017) Long short-term memory RNN for biomedical named entity recognition. BMC Bioinform 18(1):1–11
ElAbd H et al (2020) Amino acid encoding for deep learning applications. BMC Bioinform 21(1):1–14
Liu J, Gong X (2019) Attention mechanism enhanced LSTM with residual architecture and its application for protein-protein interaction residue pairs prediction. BMC Bioinform 20(1):1–11
Wang D et al (2017) MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics 33(24):3909–3916
Zhao Y, Shao J, Asmann YW (2022) Assessment and optimization of explainable machine learning models applied to transcriptomic data. Genom Proteom Bioinform 20:899–911
Souri A et al (2020) A new machine learning-based healthcare monitoring model for student’s condition diagnosis in Internet of Things environment. Soft Comput 24(22):17111–17121
D’Orazio M et al (2022) Machine learning phenomics (MLP) combining deep learning with time-lapse-microscopy for monitoring colorectal adenocarcinoma cells gene expression and drug-response. Sci Rep 12(1):1–14
Karim MR et al (2021) Deep learning-based clustering approaches for bioinformatics. Brief Bioinform 22(1):393–415
Aydin Z (2020) Performance analysis of machine learning and bioinformatics applications on high performance computing systems. Acad Platf J Eng Sci 8(1):1–14
Mohamed Shakeel P et al (2018) Maintaining security and privacy in health care system using learning based deep-Q-networks. J Med Syst 42(10):1–10
Huang L et al (2019) Patient clustering improves efficiency of federated machine learning to predict mortality and hospital stay time using distributed electronic medical records. J Biomed Inform 99:103291
Wang X, Jiang X, Vaidya J (2021) Efficient verification for outsourced genome-wide association studies. J Biomed Inform 117:103714
Cui J et al (2021) FeARH: Federated machine learning with anonymous random hybridization on electronic medical records. J Biomed Inform 117:103735
Giansanti V et al (2019) Comparing deep and machine learning approaches in bioinformatics: a miRNA-target prediction case study. In: Computational science–ICCS 2019: 19th international conference, Faro, Portugal, June 12–14, 2019, proceedings, part III, vol 19, 2019. Springer
Lyu C et al (2017) Long short-term memory RNN for biomedical named entity recognition. BMC Bioinform 18:1–11
ElAbd H et al (2020) Amino acid encoding for deep learning applications. BMC Bioinform 21:1–14
Liu J, Gong X (2019) Attention mechanism enhanced LSTM with residual architecture and its application for protein–protein interaction residue pairs prediction. BMC Bioinform 20:1–11
Mohamed Shakeel P et al (2018) Maintaining security and privacy in health care system using learning based deep-Q-networks. J Med Syst 42:1–10
Sarbaz M et al (2022) Adaptive optimal control of chaotic system using backstepping neural network concept. In: 2022 8th international conference on control, instrumentation and automation (ICCIA). 2022. IEEE
Bagheri M et al (2020) Data conditioning and forecasting methodology using machine learning on production data for a well pad. In: Offshore technology conference. 2020. OTC
Soleimani R, Lobaton E (2022) Enhancing inference on physiological and kinematic periodic signals via phase-based interpretability and multi-task learning. Information 13(7):326
Mirzaeibonehkhater M (2018) Developing a dynamic recommendation system for personalizing educational content within an e-learning network. 2018: Purdue University
Morteza A et al (2023) Deep learning hyperparameter optimization: application to electricity and heat demand prediction for buildings. Energy Build 289:113036
Webber J et al (2017) Study on idle slot availability prediction for WLAN using a probabilistic neural network. In: 2017 23rd Asia-Pacific conference on communications (APCC). 2017. IEEE
Webber J et al (2022) Improved human activity recognition using majority combining of reduced-complexity sensor branch classifiers. Electronics 11(3):392
Gera T et al (2021) Dominant feature selection and machine learning-based hybrid approach to analyze android ransomware. Secur Commun Netw 2021:1–22
Bukhari SNH, Webber J, Mehbodniya A (2022) Decision tree based ensemble machine learning model for the prediction of Zika virus T-cell epitopes as potential vaccine candidates. Sci Rep 12(1):7810
Heidari A et al (2023) Machine learning applications in internet-of-drones: systematic review, recent deployments, and open issues. ACM Comput Surv 55(12):1–45
Singh R et al (2022) Analysis of network slicing for management of 5G networks using machine learning techniques. Wirel Commun Mobile Comput 2022:9169568
He P et al (2022) Towards green smart cities using Internet of Things and optimization algorithms: a systematic and bibliometric review. Sustain Comput Inform Syst 36:100822
Sadi M et al (2022) Special session: on the reliability of conventional and quantum neural network hardware. In: 2022 IEEE 40th VLSI test symposium (VTS). 2022. IEEE
Moradi M, Weng Y, Lai Y-C (2022) Defending smart electrical power grids against cyberattacks with deep Q-learning. P R X Energy 1:033005
Zhai Z-M et al (2023) Detecting weak physical signal from noise: a machine-learning approach with applications to magnetic-anomaly-guided navigation. Phys Rev Appl 19(3):034030
Li Z, Han C, Coit DW (2023) System reliability models with dependent degradation processes. In: Advances in reliability and maintainability methods and engineering applications: essays in honor of professor Hong-Zhong Huang on his 60th birthday. 2023. Springer, pp 475–497
Zhang Y et al (2019) Fault diagnosis strategy of CNC machine tools based on cascading failure. J Intell Manuf 30:2193–2202
Shen G, Zeng W, Han C, Liu P, Zhang Y (2017) Determination of the average maintenance time of CNC machine tools based on type II failure correlation. Eksploatacja i Niezawodność 19(4)
Shen G et al (2018) Fault analysis of machine tools based on grey relational analysis and main factor analysis. J Phys Conf Ser. IOP Publishing
Han C, Fu X (2023) Challenge and opportunity: deep learning-based stock price prediction by using Bi-directional LSTM model. Front Bus Econ Manag 8(2):51–54
Darbandi M (2017) Proposing new intelligent system for suggesting better service providers in cloud computing based on Kalman filtering. Int J Technol Innov Res 24(1):1–9
Dehghani F, Larijani A (2023) Average portfolio optimization using multi-layer neural networks with risk consideration. Available at SSRN, 2023
Rezaei M, Rastgoo R, Athitsos V (2023) TriHorn-Net: a model for accurate depth-based 3D hand pose estimation. Expert Syst Appl 223:119922
Ahmadi SS, Khotanlou H (2022) A hybrid of inference and stacked classifiers to indoor scenes classification of rgb-d images. In: 2022 International conference on machine vision and image processing (MVIP). 2022. IEEE
Mirzapour O, Arpanahi SK (2017) Photovoltaic parameter estimation using heuristic optimization. In: 2017 IEEE 4th international conference on knowledge-based engineering and innovation (KBEI). 2017. IEEE
Khorshidi M, Ameri M, Goli A (2023) Cracking performance evaluation and modelling of RAP mixtures containing different recycled materials using deep neural network model. Road Mater Pavement Des 1–20
Rastegar RM et al (2022) From evidence to assessment: DEVELOPING a scenario-based computational design algorithm to support informed decision-making in primary care clinic design workflow. Int J Archit Comput 20(3):567–586
Esmaeili N, Bamdad Soofi J (2022) Expounding the knowledge conversion processes within the occupational safety and health management system (OSH-MS) using concept mapping. Int J Occup Saf Ergon 28(2):1000–1015
Akyash M, Mohammadzade H, Behroozi H (2021) Dtw-merge: a novel data augmentation technique for time series classification. arXiv preprint arXiv:2103.01119
Darbandi M (2017) Proposing new intelligence algorithm for suggesting better services to cloud users based on Kalman filtering. J Comput Sci Appl 5(1):11–16
Darbandi M (2017) Kalman filtering for estimation and prediction servers with lower traffic loads for transferring high-level processes in cloud computing. Int J Technol Innov Res 23(1):10–20
Liu H et al (2023) MEMS piezoelectric resonant microphone array for lung sound classification. J Micromech Microeng 33(4):044003
Loghmani N, Moqadam R, Allahverdy A (2022) Brain tumor segmentation using multimodal mri and convolutional neural network. In: 2022 30th international conference on electrical engineering (ICEE). 2022. IEEE
Niknejad N, Caro JL, Bidese-Puhl R, Bao Y, Staiger EA (2023) Equine kinematic gait analysis using stereo videography and deep learning: stride length and stance duration estimation. J ASABE 66(4):865–877
Amiri Z et al (2023) Resilient and dependability management in distributed environments: a systematic and comprehensive literature review. Clust Comput 26(2):1565–1600
Zeng Q et al (2020) Hyperpolarized Xe NMR signal advancement by metal-organic framework entrapment in aqueous solution. Proc Natl Acad Sci 117(30):17558–17563
Liu N et al (2021) An eyelid parameters auto-measuring method based on 3D scanning. Displays 69:102063
Li C et al (2021) Long noncoding RNA p21 enhances autophagy to alleviate endothelial progenitor cells damage and promote endothelial repair in hypertension through SESN2/AMPK/TSC2 pathway. Pharmacol Res 173:105920
Li B et al (2022) Dynamic event-triggered security control for networked control systems with cyber-attacks: a model predictive control approach. Inf Sci 612:384–398
Li H, Peng R, Wang Z-A (2018) On a diffusive susceptible-infected-susceptible epidemic model with mass action mechanism and birth-death effect: analysis, simulations, and comparison with other mechanisms. SIAM J Appl Math 78(4):2129–2153
Amiri Z et al (2023) The personal health applications of machine learning techniques in the internet of behaviors. Sustainability 15(16):12406
Zhu Y et al (2021) Deep learning-based predictive identification of neural stem cell differentiation. Nat Commun 12(1):2614
Yang S et al (2022) Dual-level representation enhancement on characteristic and context for image-text retrieval. IEEE Trans Circuits Syst Video Technol 32(11):8037–8050
Yan L et al (2023) Multi-feature fusing local directional ternary pattern for facial expressions signal recognition based on video communication system. Alex Eng J 63:307–320
Dai X et al (2022) Task co-offloading for d2d-assisted mobile edge computing in industrial internet of things. IEEE Trans Industr Inf 19(1):480–490
Yan L et al (2021) Method of reaching consensus on probability of food safety based on the integration of finite credible data on block chain. IEEE access 9:123764–123776
Jiang H et al (2020) An energy-efficient framework for internet of things underlaying heterogeneous small cell networks. IEEE Trans Mob Comput 21(1):31–43
Sun L, Zhang M, Wang B, Tiwari P (2023) Few-shot class-incremental learning for medical time series classification. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2023.3247861
Gao Z, Pan X, Shao J, Jiang X, Su Z, Jin K, Ye J (2023) Automatic interpretation and clinical evaluation for fundus fluorescein angiography images of diabetic retinopathy patients by deep learning. Br J Ophthalmol 107(12):1852–1858
Wang H et al (2022) Transcranial alternating current stimulation for treating depression: a randomized controlled trial. Brain 145(1):83–91
Luan D et al (2022) Robust two-stage location allocation for emergency temporary blood supply in postdisaster. Discret Dyn Nat Soc 2022:1–20
Chen G et al (2022) Continuance intention mechanism of middle school student users on online learning platform based on qualitative comparative analysis method. Math Probl Eng 2022:1–12
Cui G et al (2013) Synthesis and characterization of Eu (III) complexes of modified cellulose and poly (N-isopropylacrylamide). Carbohyd Polym 94(1):77–81
Cheng B et al (2016) Situation-aware IoT service coordination using the event-driven SOA paradigm. IEEE Trans Netw Serv Manag 13(2):349–361
Cheng B et al (2017) Situation-aware dynamic service coordination in an IoT environment. IEEE/ACM Trans Netw 25(4):2082–2095
Zhuang Y, Jiang N, Xu Y (2022) Progressive distributed and parallel similarity retrieval of large CT image sequences in mobile telemedicine networks. Wirel Commun Mob Comput 2022:1–13
Tang Y et al (2021) An improved method for soft tissue modeling. Biomed Signal Process Control 65:102367
Zhang Z et al (2022) Endoscope image mosaic based on pyramid ORB. Biomed Signal Process Control 71:103261
Lu S et al (2023) Iterative reconstruction of low-dose CT based on differential sparse. Biomed Signal Process Control 79:104204
Lu S et al (2023) Soft tissue feature tracking based on deepmatching network. CMES Comput Model Eng Sci 136(1):363
Liu M et al (2023) Three-dimensional modeling of heart soft tissue motion. Appl Sci 13(4):2493
Heidari A et al (2023) A hybrid approach for latency and battery lifetime optimization in IoT devices through offloading and CNN learning. Sustain Comput Inform Syst 39:100899
Heidari A, Jafari Navimipour N, Unal M (2022) The history of computing in Iran (Persia)—since the achaemenid empire. Technologies 10(4):94
Ahmadpour S-S, Heidari A, Navimpour NJ, Asadi M-A, Yalcin S (2023) An efficient design of multiplier for using in nano-scale IoT systems using atomic silicon. IEEE Internet Things J 10(16):14908–14909. https://doi.org/10.1109/JIOT.2023.3267165
Amiri Z, Heidari A, Navimipour NJ et al (2023) Adventures in data analysis: a systematic review of deep learning techniques for pattern recognition in cyber-physical-social systems. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-16382-x
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amiri, Z., Heidari, A., Navimipour, N.J. et al. The deep learning applications in IoT-based bio- and medical informatics: a systematic literature review. Neural Comput & Applic 36, 5757–5797 (2024). https://doi.org/10.1007/s00521-023-09366-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-09366-3