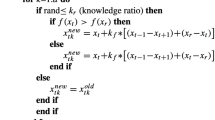Abstract
Feature selection (FS) is one of the basic data preprocessing steps in data mining and machine learning. It is used to reduce feature size and increase model generalization. In addition to minimizing feature dimensionality, it also enhances classification accuracy and reduces model complexity, which are essential in several applications. Traditional methods for feature selection often fail in the optimal global solution due to the large search space. Many hybrid techniques have been proposed depending on merging several search strategies which have been used individually as a solution to the FS problem. This study proposes a modified hunger games search algorithm (mHGS), for solving optimization and FS problems. The main advantages of the proposed mHGS are to resolve the following drawbacks that have been raised in the original HGS; (1) avoiding the local search, (2) solving the problem of premature convergence, and (3) balancing between the exploitation and exploration phases. The mHGS has been evaluated by using the IEEE Congress on Evolutionary Computation 2020 (CEC’20) for optimization test and ten medical and chemical datasets. The data have dimensions up to 20000 features or more. The results of the proposed algorithm have been compared to a variety of well-known optimization methods, including improved multi-operator differential evolution algorithm (IMODE), gravitational search algorithm, grey wolf optimization, Harris Hawks optimization, whale optimization algorithm, slime mould algorithm and hunger search games search. The experimental results suggest that the proposed mHGS can generate effective search results without increasing the computational cost and improving the convergence speed. It has also improved the SVM classification performance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Optimization is a procedure for maximizing or minimizing an objective function or multiple objectives [1]. Many problems can be handled by employing several optimization techniques to find the optimum solution. Optimization is used to create optimal paths in real life. Metaheuristics are useful tools that provide different ways of creating effective optimization algorithms. Although the exact solution is not offered, the algorithm can provide the best possible solutions. Feature selection (FS) is one of the most important preprocessing steps in data mining and pattern recognition. Its aim is to filter features and select a subset from a given training dataset. One of the benefits of FS is that it reduces the training time for model creation, eliminates overfitting, and improves the generalization of the models for a variety of datasets, such as biomedical signal processing, medical images, DNA microarray data, chemical data, and drug development. The feature dimensions of the data acquired from multiple medical sources are incredibly high. Relevant literature has demonstrated that applying FS to various medical domain data has a considerable effect on the results. There are various methods for FS used in machine learning (ML) and data mining [2]. They can be divided into filtering, wrapper, and embedding (hybrid method).
First, filtering techniques evaluate each feature subset using an objective function based on target relevance or feature correlation. Second, wrapper approaches explore a feature subset based on a predefined classifier performance score. Third, hybrid method is the combination of filter and wrapper method that is built by the algorithm has built-in FS methods. Example of this method ridge regression has penalization inbuilt. FS can be considered an NP hard problem as there are many possible solutions, especially for large feature space [3]. A binary version of several metaheuristic algorithms (MAs), such as wrapper techniques for FS, has been proposed to provide a suitable solution. Some MAs such as genetic algorithm (GA) [4], particle swarm optimization (PSO) [5], bee colony optimization (BCO) [6], cuckoo search (CS) [7], grey wolf optimizer (GWO) [8], improved multi-operator differential evolution algorithm (IMODE) [9], gravitational search algorithm (GSA) [10], Harris’ Hawks optimization (HHO) [1], whale optimization algorithm (WOA) [11], and slime mould algorithm (SMA) [12] have been applied for FS and support vector machine (SVM) kernel parameters are optimized simultaneously [13]. FS has been widely used in sequence analysis for Bioinformatics. Content and signal analysis is two sorts of challenges that can be solved using FS. The content analysis examines a sequence’s general properties, such as its propensity to code for proteins or its ability to perform a certain biological function. In contrast, the signal analysis identifies key motifs in the sequence as regulatory elements or gene structural elements.
This study proposes a modified hunger games search (mHGS) hybrid metaheuristic algorithm to solve the problems of classical HGS optimization algorithm. It simulates the hungry behaviour in animals. For FS, mHGS algorithm performance was assessed by well-known benchmark test functions and a set of well-known medical datasets. The fitness function is based on a straight-forward understanding of hunger as a critical biological drive. The fitness value for hunger is higher than other compared algorithms. Other hunger games can alter their initial positions based on the fittest hunger game. These behaviours can be related to the goal function to be optimized. The mHGS effectiveness has been tested on the complex CEC’20 benchmark functions and several biomedical datasets, and compared with several counterpart MHAs including IMODE, GSA, GWO, HHO, WOA, SMA, and original HGS algorithm [14]. The experimental results proved that the proposed mHGS has a stronger search capability than the basic HGS, and some state-of-the-art MH methods.
Motivation Despite eminent applications, the hunger games search (HGS) has attracted the attention of many researchers—the method has reached more than 200 citations in about a year. HGS is still attributed for its slow convergence and stagnancy issues when employed on high-dimensional problems [15]. It sometimes generates low-diversified solutions towards the end of iterations, which causes a situation for the search agents trapped in local optima. Additionally, its exploration phase is highly dominant with extraordinary randomization that seems clueless search mechanism. The promising outcomes of several HGS-based hybrid approaches proposed in the literature, especially HGS, have been carried out with chaotic maps in three alternative scenarios [16]. Additionally, HGS is merged with the food-searching techniques of the whale optimization algorithm (WOA) for global optimization [17]. In the same context, in order to identify unknown parameters of solar photovoltaic (PV) systems, the Nelder-Mead simplex method (NMs) is injected in HGS to increase the population diversity [18].
In this study, the proposed mHGS algorithm integrates the efficacy of fuzzy into HGS’s exploration phase. Moreover, additional modifications are also proposed to select animals. The enhanced HGS variant is not only tested on some classification datasets (with feature-size greater than 15000) for feature selection problem, also it is evaluated on hard numerical optimization problems presented in CEC’20 test suite which is well-known in optimization community for its difficult search space. The outcome of the simulations performed in this research reveals superiority of the proposed approach when compared with the conventional HGS and other optimization algorithms, as well as, several other counterparts introduced recently; however, it lags superiority on state-of-the-art methods like improved multi-operator differential evolution algorithm (IMODE) [9], gravitational search algorithm (GSA) [10], grey wolf optimization (GWO) [8], Harris Hawks optimization (HHO) [1], whale optimization algorithm (WOA) [11], and slime mould algorithm (SMA) [12]. The proposed mHGS maintains trade-off balance between exploration and exploitation, convergence speed, and better global search ability. The argument is well supported with various statistical measurements and evaluation metrics later in this paper.
In summary, the main contributions of this paper are as follows:
-
The traditional HGS is enhanced by adding the following mechanism: Fuzzy logic-based mutation for control parameters strategy, balancing exploration/exploitation strategy, and population reduction strategy.
-
mHGS is proposed as an alternate feature selection approach.
-
mHGS is proposed to improve its local search capability and solve the problem of premature convergence.
-
The proposed mHGS algorithm achieved superior results compared to its counterparts.
-
Various metrics from statistical to qualitative analyses assess the performance of the proposed mHGS algorithm.
The following is how the rest of the paper is organized: Sect. 2 presents the related work, highlighting several recent related research. Preliminaries on QSAR approach, hunger games search (HGS), and support vector machine (SVM) are introduced in Sect. 3. Section 4 discusses the proposed mHGS algorithm including the fuzzy logic basics, fuzzy logic-based mutation, membership function formation (MFs), centroid-based fuzzy mutation, fitness function (fobj), pseudo-code of mHGS algorithm, and mHGS development phases. Section 5 introduces two experimental series, including the CEC’20 benchmark functions, feature selection (FS) and discussion. Finally, Sect. 6 shows the conclusions and future work.
2 Related research
Here, we highlight some of the important research in line with the proposed work. In [19], for creating a medical molecule, diagnosing several diseases, or determining the optimal drug, so it is necessary to collect relevant data. There are many ways of collecting medical or chemical data. First, for medical molecules, protein bank and ZINC databases are used to select the suitable crystal of protein structure. One of the most efficient methods for making drugs from chemical data is computer-aided drug design (CADD). In [20], CADD is used to identify drug design which can be classified into quantitative structure-activity relationship (QSAR) and docking. The binding between protein and ligand is docking. Many chemical compounds can be extracted from the Pubchem website.
In [21], proteins and ligands are tied with each other, so ligands must be separated from the protein. The best drug depends on the best ligands with less energy. Pymol software is used for the separation operation. AutoDocks software is used to calculate the energy for several ligands [22].
In [23], multiple tasks have been proposed with a drug review dataset. Sentiment analysis is used to predict user sentiments about medicine side effects and effectiveness based on user reviews. The transferability of trained classification models among domains has been investigated to overcome several challenges for lacking annotated data. The transfer learning approaches have been proposed to indicate the similarities across different domains. For all prediction tasks, classification-based sentiment analyses, as an n-grams approach, are applied to indicate several user reviews. The classification model for this dataset achieves 92.24% accuracy. A method based on biclustering has been discussed in [24] to reduce the molecular descriptors number to predict chemical compounds biodegradation. Several classifiers were used to assess the biodegradation task. For the QSAR biodegradation data, the testing results indicate that random forest is the best classifier, with an accuracy of 88.81% and only 19 MD.
In [25, 26], artificial intelligence (AI) has made significant progress, allowing the development of various horizons for QSAR modelling based on machine learning. In [27], some authors proposed combining artificial neural networks (ANN) and support vector machines (SVM) for QSAR modelling, with PCA used to minimize data dimensionality. The performance is measured on the QSAR Biodegradation dataset, with an accuracy 82%. In [28], silico models have been described to identify the organic AR modulators. ML methods predicted AR binding based on the tree classification model as k-nearest neighbour (k-NN), RF, and naive Bayes. The models achieved robust and reliable predictions.
In [29], the optimum treatment for the two most frequent forms of warts, plantar and common has been determined in response to two of the best treatment modalities such as immunotherapy and cryotherapy). The treatment approach was chosen at random using fuzzy rule-based inference mechanism to forecast treatment technique response. The percentages for immunotherapy and cryotherapy were 83.33% and 80.7%, respectively. Moreover, such expert systems reduce treatment costs, save time for patients, and also improve treatment quality. In [30], two peptide datasets have been used; one for lung cancer cells and the other for breast cancer cells. ANN has also been used for recognizing the peptides inducing breast/lung cancer death. Fourteen peptides from 1000 denovo designs were selected for in vitro testing and production on breast cancer. In [31], it has been indicated as the most frequent cause of dementia in the world, Alzheimer’s disease. Due to the timeliness of diagnosis and the ageing of the population, it has begun to outrun. Due to a lack of sensitivity and precision, case classification, magnetic resonance imaging, and neuropsychological testing are ineffective. The convolutional neural network has been used to construct a framework to indicate many Alzheimer’s disease features.
In [32], FS is divided into four steps: (1) select the appropriate features, (2) analyse the subset using various metrics, (3) locate other sets, and (4) feature validation. In [33, 34], it has been shown that wrapper-based solutions outperform filter approaches in terms of results. The wrapper approach is more time-consuming, yet yields precise results. FS may be tweaked to be more efficient by identifying the best subset of attributes to tackle a range of problems. MHAs have many advantages.
The authors in [35] suggest that the wrapper-based techniques have attracted significant attention due to the use of learning algorithms which influence the selection of significant features. In [36], the MHA-based approach SSA using wrapper FS has been presented to estimate chemical compound activity by determining the most suitable subset of molecular descriptors from the MAO dataset. SSA is compared with many MHAs, including the moth-flame optimization algorithm (MFO), grasshopper optimization algorithm, and sine cosine algorithm. It is worth noting that SSA with the k-NN classifier had the best accuracy of 87.35% while keeping 783 chemical descriptors. In [37], two classification approaches, HHO-SVM and HHO-kNN, for drug design and discovery prediction have been proposed. Several techniques for FS are explained. In [38], the strategies for selecting medications based on their features and the importance of chemical descriptors have been presented. FS can be considered a multi-objective optimization problem by decreasing the number of the selected features and increasing the accuracy.
In [39], some authors described the FS approach used in medication development. During the FS phase, crude set-based approaches are used to identify the most discriminative features. The most discriminative traits were chosen using rough set-based approaches. Several features were selected from the feature vector at this step using three distinct rough set-based methods, such as QRFS, DMFS, and EBFS. Using these algorithms has the goal of reducing the number of features to improve classification performance and reduce classification time. In [40], the authors suggested FS techniques used in various domains, primarily to deal with data with high dimensions. Several FS solutions based on (MHAs) methods have been developed to address the FS problem and overcome the limits of classic FS approaches. Also, in [41], the authors outlined how ML system requires FS. The effectiveness of such systems heavily depends on the relevancy of features to the target. FS is an NP-hard problem because there are many alternative solutions, especially for large feature space. A novel multi-population-based PSO (MPPSO) has been presented for FS.
In [42], MHAs are used to determine important features to boost the performance of high-dimensional datasets to devise efficient knowledge extraction systems declared FS as an important preprocessing step that helps to avoid the effect of noisy, and inconsistent features on model performance. However, when used with dataset with several features these algorithms frequently suffer from a local optimally problem because of the large solution space. In [43], a unique technique for dimensionality reduction has been used to improve classification accuracy, which uses the Henry gas solubility optimization process for picking relevant features. In [44], many feature extraction approaches have been presented to test their prediction performance. During the testing step, it will be necessary to specify the facial picture view and the recorded eye-gaze locations. Table 1 depicts some published literature studies on related topics.
3 Preliminaries
The basic QSAR methodologies and structure of hunger game optimization and SVM are explained in this section.
3.1 Quantitative structure-activity relationship (QSAR)
QSAR is used to express the chemical structure and the biological activity relationship in a mathematical form. It is useful, especially when it is used to recognize the chemical compounds’ features. Many ML algorithms have been successfully used to analyse structure-activity relationship to predict whether a substance is similar to a drug-like activity. Many complicated molecular compounds can be applied to characterize variety of properties. In chemistry and pharmacology, molecular descriptors are crucial [52]. QSAR techniques are based on developing statistical models for the association between chemical structure and biological activity. The classification process has been used in the chemical-biological interaction between many biomolecules.
In QSAR studies, the next step is to develop a statistical approach using the descriptors obtained previously from several compounds. This model’s primary purpose is to predict activity for new compounds and to use it for understanding the action predicted for a specific drug. The input data accuracy, descriptors and statistical techniques selection, and produced model validation play a significant role in the quality of a QSAR model [53]. In the same context, the ANNs and SVMs are applied in the QSAR field and other molecular modelling approaches have recently attracted significant attention as key tools in drug discovery as discussed in [54, 55].
3.2 Hunger games search (HGS)
The HGS algorithm is discussed in this section, with its mathematical model [14]. The HGS simulates the hunger behaviour of selected animals. HGS fitness is determined by the approach used to track hunger behaviour as a critical homeostatic incentive. Many behaviours that generate action and choice in the animals’ lives are used to confine and understand the optimization process for new uses. The algorithm’s feature process is an adaptive weight based on the hunger concept used and built to replicate each hunger effect search stage. Most animals’ computationally used logical principles and game activities are adaptive evolutionary chances for food acquisition. The basic notion is that the proposed technique is more efficient because of its high performance, dynamic nature, and simple structure in terms of convergence and quality for acceptable solutions. HGS performance was compared with other optimization algorithms with several experimental results such as CEC’20 benchmark.
3.2.1 Approach food
Mathematical formulas express the behaviour of approach food, and the following procedures are proposed to simulate the contraction mode. The mathematical formulas Eq. (1) is as follows:
where R is within \([-a,a]\); \(r_1\) and \(r_2\) represent random numbers within [0,1]; \(W_{1}\) and \(W_{2}\) denote hunger weights; \(X_b\) denotes a random person in the population; and X(t) is an individual. The formula of Eq. (2) is as follows:
where \(i \in 1, r, \ldots , n, F(i)\) is the fitness for an individual i, whereas the best fitness value for the current iteration is denoted by BF. The hyperbolic function (Sech) is represented as (3):
3.2.2 Hunger role
Equations (5) and (6) represent the mathematical formulation of the role:
where the population of individuals is denoted by N, whereas SHungry is sum(hungry). The randomization in search is injected using random variables \(r_{3}\), \(r_{4}\) and \(r_{5}\). Equation (7) is a formulation for hungry(i):
where AllFitness(i) preserves each individual’s fitness in the current iteration.
where \(r_{6}\) is another randomization variable and F(i) indicates each individual’s fitness value. The best fitness and worst fitness values are denoted by BF and WF, whereas the upper and lower search bounds are UB and LB, respectively. Because the hunger sensation H has a lower bound, LH, it contributes to the algorithm’s optimum performance.
3.3 Support vector machine
It is a supervised learning techniques applied in classification tasks [56]. Using the core concepts for the nonlinear kernel function, the approach is used to map data from a high-dimensional space. To identify the optimum solution for separating two classes, the SVM is utilized. Regression and classification problems are solved by the SVM model. The basic principle behind SVM is that the algorithm generates a hyperplane that is used to divide data into classes. SVMs’ first major challenge is to find a dividing line (or hyperplane) between data from two classes. SVM is a classification technique that uses data to produce a line that divides the classes. It aims to optimize different margins in the closest positions, referred to as support vectors and hyperplanes. The algorithm output is an optimal hyperplane. This hyperplane is a two-dimensional line that splits a plane into two sections, each with its own class.
In the SVM result, various parameters are controlled. The arguments determined by the designer when the classifier is formed are referred to as tuning parameters. The C parameter regulates the balance between a smooth decision boundary and correctly identifying training points. If C a large value is defined, it will appropriately obtain more training points, but it will also result in more convoluted decision curves attempting to fit all of the points into the output. To create a properly balanced curve and avoid overfitting, multiple values of C were used for the same dataset. Meanwhile, \(\varGamma \) describes the range of influence of a single training session. If it has a low value, every point will have a long reach, and if it has a high value, every point will have a close reach. If \(\varGamma \) has a very high value, then the decision boundary will be based solely on the points that are extremely close to the line, essentially ignoring some of the points that are quite far from the decision boundary. This is due to the fact that the points that are closer have a greater weight. If the \(\varGamma \) value is low, even the far points gain a high weight.
SVM is a supervised learning technique used in classification [56]. Using the core concepts for the nonlinear kernel function, the svm approach is used to map data from a high-dimensional space. The SVM is utilized to identify the optimum solution for separating two classes. The SVM represents a linear model for regression and classification problems. It is an efficient method for various real situations and can solve both linear and non-linear problems. The basic principle behind SVM is that the algorithm generates a line or hyperplane that is used to divide data into classes. SVMs’ first major challenge is to find a dividing line (or hyperplane) between data from different classes. It aims to optimize different margins in the closest positions, referred to as support vectors and hyperplanes. The algorithm output is an optimal hyperplane. This hyperplane is a two-dimensional plane line that divides a plane into two sections, each class on either side. In the SVM result, various parameters are controlled. The arguments determined by the designer when the classifier is formed are referred to as tuning parameters.
In cheminformatics, SVM is one of the most widely used machine learning algorithms. The prediction of toxicity-related qualities such as mutagenic toxicity and toxicity categorization is one of the uses of SVM.
SVM algorithm is introduced in Algorithm 1, and also Fig. 1 is used for declaration.
A graphical explanation is shown in Fig. 1.

4 The proposed mHGS
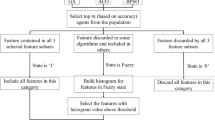
The proposed mHGS algorithm has been developed to address the HGS problem, which includes being caught in sub-optimal regions, delayed convergence, and is commended balance between global and local search, illustrated in convergence curves in Fig.4. Our enhancement strategy is based on two different testing methods,including CEC’20 and biological and chemical data with various evaluation criteria.
4.1 Fuzzy logic
The fuzzy set theory [57] was introduced in 1965. It is used increasingly and employed in several domains, including image segmentation [58], cancer classification [59], and optimization [60]. Most natural objects cannot be defined because of simple shapes. Fuzzy logic’s characteristics are based on the truth value of a variable as a real number between 0 and 1. Set theory’s fuzziness is depicted graphically for future reference. Membership functions (MFs) can be used to define it. In the universal domain, any fuzzy set Eq. (10) is a set of ordered pairs as follows:
where x is a universal set of U element knowledge ; \(\mu _{F}\) is the MF for F with values in [0, 1]. In the current domain, the existence of MF is an expertise element that can have many membership levels. Although the fuzzy set boundary is a nonzero element, it is an incomplete membership \((<1)\).
4.2 Fuzzy logic-based mutation
The fuzzy logic notion is applied to address various research challenges in industrial applications. An MF indicates the membership value for an operation. Any fuzzy set can be defined as a collection of ordered pairs in the universal domain. Here, the MF of its \(\hbox {i}{\hat{\hbox {t}}}\hbox {h}\) value is in the range [0, 1], and it is a universal set information element. Consequently, one element of information can have multiple degrees of membership in the current domain, depending on the nature of the MF. The fuzzy set’ core comprises of elements with full membership, where as support comprises of elements with nonzero membership. The fuzzy set’s boundary comprises of elements with nonzero but in complete membership.
4.3 Membership functions formation (MF)
In fuzzy logic, MF plays a crucial role in the performance of fuzzy representations in different situations [57]. To be specific, the MF shape is critical for a certain problem because it controls the fuzzy inference rules. MF can be Gaussian, triangular, trapezoidal, or take other forms with their requirement that an MF’s values are between 0 and 1. MF basically maps the given data to the necessary degree of membership. A thorough study of the underlying problem can lead to the conclusion that the MF shape is appropriate for the application under certain conditions. There could be an infinite number of ways to define fuzziness. The approach depends on the nature of the problem. In addition to determining the MF shape, determining the interval and number of MFs is critical. Therefore, to regulate the system model in temperature terms using fuzzy logic, it is necessary to identify numerous MF (high, medium, and low) membership value intervals. These variables have a substantial impact on the inference of a fuzzy logic-based system. Observing data distribution is also a significant component. The trial and error method is sometimes employed to choose a MF form. The function can use any curve, as long as it meets the efficiency, simplicity, and speed criteria. However, the MF number has a significant impact on the computational time. As a result, the best model for achieving the best system performance can be identified by varying types and MFS. Some MF concepts are literally explored, such as fuzzy logic as a universal approximate or constrained interpolations, which are suitable for MF with finite membership values [61].
The main goal is to separate the 0-1 modelling, which may be accomplished with a triangular MF; however, a more complex situation may necessitate using a specific MF type. A high-fidelity intuition based on sufficient experience can provide a good solution for making the optimal choice. To find memberships for fuzzy variables, MH optimization and evolutionary optimization techniques are used, e.g. NNs and GA. The fuzzy logic approach was used to determine the particle’s performed mutation probability. At any given time, a particle’s mutation probability is not totally certain or uncertain; instead, the membership value provides the mutation probability.
4.4 Centroid-based fuzzy mutation
This concept is the development of a mutation that aids the population particles in determining when to drift, allowing for premature convergence. There are two crucial parameters to consider with a particle mutation treatment: particles distance from other particles and particles history. As the particle distance from the population centroid is checked, the accuracy of the particles changes. When particles are closer to one another, early convergence is possible. Meanwhile, to avoid this problem, the mutation method is used to separate the particles. This can assist many particles in overcoming the local solution. Estimating population distance from the centroid of other populations is more appropriate than calculating particle distance from other particles. If the distance between populations is small, it means that the populations are close to each other.
The population distance from the centroid is inversely proportional to the mutation probability. In some cases, it is possible that a population lives at the centroid. When the distance is 0, there is an infinite mutation chance, which is undesirable. Therefore, we add one to the distance to ensure that this scenario does not occur. As a result, from Eq. (11) declares the contribution of distance to mutation probability as follows:
where dist is how much a particle is far from the centroid, P\(_{d}\) is approximate contribution, is the distance contribution to the mutation probability. In a similar way to distance, population history may play a role in mutation probability. If the best solution is reached, the frequency of iterations declared by the population will be changed. The best solutions are still explored and trying to find a better solution. But, if the global solution stays static over iterations, this indicates that these solutions are trapped in local solutions and cannot be searched in several parts of the search space. Therefore, mutations in the populations are important to provide some perturbation between them, thereby helping several populations to avoid local optima and find solutions in global optima. If the time for the global best populations is always expanding, raising the mutation probability is also required. This historical information contribution is calculated by (\(P_{c}\)). The global population has remained unaffected by using the iteration number unchanged for mutation probability. \(\alpha =4\) and \(\beta =5\), Following these values, the probability of mutation increases as the value of increased.
where a = 0.5 and b = 0.5, the tan function returns a number between [1, 1] and [0.5, 0.5] that, when multiplied by b, is constrained to [0.5, 0.5]. As a result, the final value is in the range [0, 1]. Equivalent Eq. (12) is used to combine two contributions. Where the parameters \(\alpha \) and \(\beta \) gave the distance and background contributions equal weight. The values of \(\alpha \) and \(\beta \) were set to 0.6 and 0.4, respectively. As an example, distance has become increasingly important throughout time. Other particles change, even if the best does not. In this situation, some particles may not be trapped, but the solution will consider the potential for convergence. To avoid this, Eq. (12) assigns a lower weight to history.
Mutation probability Eq. (13) for the \(\hbox {i}{\hat{\hbox {t}}}\hbox {h}\) particle in a population is declared by \(P_{i}\). If \(P_{i}\) is greater than a randomly value generated of particle i, the particle is muted, otherwise mutation does not occur. The mutation is done by the two Eqs. (14, 15).
(1) where \(\varDelta p\) indicates the change in the particle’s value in the jth dimension, this range denotes the difference between the benchmark function scope’s upper and lower limits, count denotes the current iteration number, iter denotes the total number of iterations to be performed, and \(P_{i j}\) denotes the value of the ith particle in the jth of the entire population dimension. The value of \(\varDelta q\) lowers as the points converge, allowing for fewer disruptions. To ensure that the disturbance in an agent’s motion is minimized, Eq. (15), \(\varDelta \) is controlled. Despite the fact that there are multiple assumptions in this scenario, the fuzzy mutant forms of the HGS algorithm often have a higher possibility of avoiding the convergence problems.
4.5 Fitness function (fobj)
This section is used to explain the mHGS fitness function (fobj), which is formulated as:
where R stands for the error rate, C stands for the summation of features in the dataset, \(\alpha \) and \(\beta \) stand for the classification quality importance (as calculated by the classifiers) and subset length, respectively. The range of \(\alpha \) is specified as [0, 1]. G stands for the classifier’s group column, and T stands for the condition that each method is compared to the fitness function. To maximize the solution, fobj must be greater than T.
Algorithm 2 describes the steps in the mHGS-SVM algorithm optimization process. In addition, a flowchart depicting the detailed steps of the mHGS procedures is shown in Fig. 2.

4.6 mHGS development phase
Initialization stage The proposed mHGS algorithm starts the optimization process randomly initialize the agents’ population using a uniform random distribution as follows:
where \(X_{i}^{\text{ initial }}\) is the random initialized \({i}{\hat{\hbox {t}}}\hbox {h}\) solution vector, UB, LB are the upper and lower bounds, respectively, n is the population size ; \({\text {rand}}_{i} \in [0,1]\) is a random value. The fitness function (fobj ) is calculated using Eq. (16).
Population reduction In population-based algorithms, the number of search agents in the population NP plays an essential role in adjusting the algorithm convergence rate. Further explanation: Small population agents can converge quickly but; however, they improve the probability obtaining a local optimum. The population with a large number of agents converges more slowly but provides a better exploration of the search space. The proposed mHGS applies the population reduction approach of linear reduction as follows:
where NPmax is the initial population size (NP); NPmin is the specified minimum population size ; NPmin \(=30\) in this study; FE is the current function evaluation; MAX_FE \(=45000\) is the maximum number of function evaluations.
Solution step Solutions are proposed, and new solutions are proposed using Eqs. (1, 2) as illustrated in Algorithm 2. Applying the fuzzy mutation to improve the search space by exploring several new regions to identify the best candidate solution using Eqs. (14), (15), which helps in improving the diversity of algorithms, and avoid local solutions and comparing between new and previous solution. The best new solution is used for the update using Eq. (6) for hunger and Eq. (7) for calculating the hunger sensation that controls the algorithm performance. Algorithm 2 declares how the best solution is proposed for calculating the objective fitness for several new populations. This process is repeated until the stopping condition is achieved.
Termination step The proposed mHGS algorithm is repeated until the stopping criteria are met, resulting in the best candidate solution. The steps of the proposed mHGS algorithm are illustrated in Figure 2, and the pseudo-code is presented in Algorithm 2.
Classification phase The best-proposed solution selected in the previous phase is obtained using the mHGS method. In Xbest, the features retrieved from the original dataset are equal to one. The SVM classification approach is implemented. The dataset is split into two sets: 90% train and 10% test. It is worth noting that all experiments were conducted 30 independent times to get the best results.
5 Experimental results and simulations
The CEC’20 benchmark functions and FS dataset are applied for evaluating the proposed mHGS including several testing such as statistical results and qualitative metrics. For fair evaluation, the suggested mHGS results were compared with other seven MAs as (IMODE), (GSA), (GWO), (HHO), (WOA), (SMA) and original HGS algorithm. Table 2 shows the parameter settings for all the compared algorithms. To generate results, all of the algorithms were checked using MATLAB programming language. For FS evaluation, all the compared algorithms were hybridized with SVM as a classifier. A total of ten chemical datasets were used. function evaluations (FEs) were utilized each comparative algorithm 30 times with 30 agents.
5.1 Parameter settings and evaluation metrics
Many compared algorithms are used in our experiment to evaluate our enhancement method. Parameter settings have a main role in controlling several conditions in our experiment. All parameters for a fair optimization experiment are defined in Table 2.
The following measurements are utilized to validate and evaluate the proposed method depended on the best fitness value fobj obtained at run i:
-
1.
The average of the fitness function values produced by running method M for times is the mean. The mean fitness function can be calculated using the following equation:
$$\begin{aligned} {\hbox {Mean}} = \frac{ \sum _{i=1}^{M} {{\hbox {fobj}}(i)} }{M} \end{aligned}$$(21) -
2.
The maximum value of the fitness function obtained by running the algorithm M times refers to the best fitness function. The value of the best fitness function can be calculated as follows:
$$\begin{aligned} {\hbox {Best}} = \max _{i=1}^{M} {{\hbox {fobj}}(i)} \end{aligned}$$(22) -
3.
The worst fitness function is the fitness function with the minimum value produced by performing the algorithm M times. The value of the worst fitness function can be computed by
$$\begin{aligned} {\hbox {Worst}} = \min _{i=1}^{M} {{\hbox {fobj}}(i)} \end{aligned}$$(23) -
4.
Standard deviation is used to measure the fluctuation of the fitness function value obtained from M times of the running algorithm (STD). STD is an indication used to measure the stability and robustness of the algorithm. Higher standard deviation values suggest that the algorithm wanders, but a smaller number shows that the method converges for the same value in the majority of iterations. Using the formula below, the standard deviation can be calculated:
$$\begin{aligned} {\hbox {STD}} = \sqrt{\frac{1}{M-1}\Sigma _{{i=1}}^{M} ({\hbox {fobj}}(i)-{\hbox {mean}}) ^2} \end{aligned}$$(24)
5.2 Experimental series 1: statistical results for CEC’20
To assess the proposed mHGS approach, the IEEE Congress on Evolutionary Computation 2020 (CEC’20) [62] was selected for evaluating its performance while solving different types of suite objective functions. Statistic methods are applied to algorithms that use CEC’20 functions to indicate which algorithm is the best.
5.2.1 CEC’20 benchmark functions description
The test data for testing the performance of the suggested algorithms were taken from the IEEE Congress on Evolutionary Computation (CEC) [62]. Ten test functions were included in the CEC’20 benchmark functions, containing unimodal, multimodal, hybrid, and composition functions.
5.2.2 Parameter space
Figure 3 declares a 2D visualization of the CEC’20 functions to help understanding the differences and each problem nature.
5.2.3 Statistical results analysis
Table 3 reports the statistical results such as mean and STD for the proposed algorithm and other compared algorithms for each CEC’20 benchmark function with 10-dimension; the best results (minimum values). The suggested approach outperformed previous algorithms in solving most of the CEC’20 benchmark functions in terms of mean and standard deviation. Furthermore, the proposed mHGS was ranked top in the Friedman mean rank-sum test.
5.2.4 Convergence behaviour analysis
The mHGS algorithm and other algorithm performance can be explained with convergence curves and counterparts with the CEC’20 functions shown in Fig. 4. All the results of the functions for the proposed algorithm reached a stable point, indicating that, it converges. Fast convergence indicates the optimal solution. Thus, the suggested mHGS method is a promising optimization to solve several problems that need fast computing, such as online optimization problems.
5.2.5 Boxplot behaviour analysis
Boxplots are used to show data distribution characteristics. The distribution results are related to several local minima of class functions, as shown in Fig. 5. Boxplots are effective for presenting data distributions in quartiles. The algorithm’s minimum and maximum data points, which are the whisker’s edges, are the algorithm’s lowest and highest data points. High level of data agreement is declared by a narrow boxplot Fig. 5. Boxplot shows the results for ten functions Dim \(=10\). The mHGS algorithm produces the best results when compared to other algorithms.
5.2.6 Qualitative metrics analysis
Particle behaviour monitoring, or search agents, for instance, can propose more knowledge about algorithm convergence and the optimization search process. The mHGS algorithm qualitative analysis is indicated in Fig. 6. The agent’s behaviours are shown in Fig. 6, which illustrates the functions in two dimensions (3D), search history, average fitness history, and convergence curves.
These points discuss the qualitative analysis
-
In terms of domain’s topology—functions in 3D views The function in 3D space is indicated in the first column of Fig. 6. The functions have distinct topologies, which aids in deciding which type or shape of function the algorithm performs best.
-
For the search history The search history of agents is shown in the second column of Fig. 6 from the first to the last iteration. The search space is represented by counter lines, with the gradient from blue to red lines indicating a higher fitness value. The suggested mHGS can locate the locations with the lowest fitness values for particular functions, according to search history.
-
In terms of average fitness history The average fitness history is shown in the third column of Fig. 6, i.e. the fitness value averages as a function of the iteration number. This average reveals the agents’ overall behaviour as well as their contribution in the optimization process. The population improves as the history curves diminish. This continuous improvement demonstrates a collaborative searching behaviour and backs up the efficacy of particle law updates.
5.3 Experimental series 2: applying mHGS for FS
5.3.1 Data description
We have used several datasets that was collected from Machine Learning Repository and kaggle websites but only MAO data set taken from GREYC’s Chemistry dataset.Footnote 1Footnote 2Footnote 3Monoamine Oxidase (MAO) The dataset is provided by an enzyme that is widely distributed in the major tissues. It has the ability to catalyse the inactivation and oxidation of monoamine neurotransmitters. The GREYC Chemistry dataset provided the data for this dataset. This data set is taken from https://brunl01.users.greyc.fr/CHEMISTRY/#MAO. Thus, MOA is transferred to SMILES (Simplified molecu//-lar-input lineen try system) styles using open babel software [63]. Then, the molecular descriptors (MD) are determined using E-dragon [64]. It has 1665 features (MD) with 68 compounds divided into two classes.
QSAR Biodegradation This dataset has 41 features (molecular descriptors) that are used to classify 1055 chemical compounds. These data are used to determine between two chemical classes, with 356 readily biodegradable samples and 699 not readily biodegradable patterns. Furthermore, this information can be used in the building of QSARs to indicate the relationship between molecular biodegradation and chemical design. It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/QSAR+biodegradation).
Drug Review This dataset includes patient reviews for specific medicines as well as diseases that are related to them. It has ten patients who have given it a rating that reflects overall patient satisfaction. Crawling online pharmaceutical review sites yielded the information. The goal was to learn something new. Splitting this data into a train (75%) and a test (25%) yields the greatest results (25%). It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/Drug+Review+Dataset+%28Drugs.com%29).
Drug consumption There are 1885 responders in the database. All input attributes are categorical at first and are then quantified. Participants were also asked about their use of 18 legal and illegal drugs, including alcohol, amphetamines, benzodiazepine, cannabis, chocolate, cocaine, caffeine, crack, ecstasy, heroin, ketamine, legal highs, LSD, methadone, mushrooms, nicotine, and volatile substance abuse, as well as one fictitious drug to identify over-claimers. It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/Drug+consumption+%28quantified%29).
QSAR androgen receptor Using various machine learning methods, this dataset was used to create classification QSAR models for the discrimination of binder/positive (199) and non-binder/negative (1488) molecules. The following reference provides more information: Machine Learning Consensus to Predict the Binding to the Androgen Receptor within the CoMPARA Project, 59, 1839-1848, Journal of Chemical Information and Modeling. The Milano Chemometrics and QSAR Research Group (Universit degli Studi Milano-Bicocca, Milano, Italy) calculated attributes (molecular fingerprints) on a set of chemicals provided by the National Center of Computational Toxicology at the US Environmental Protection Agency as part of the CoMPARA collaborative modelling project, which aimed to develop QSAR models to identify binders to the Androgen Receptor. It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/QSAR+androgen+receptor).
Immunotherapy This dataset contains 90 instances of wart treatment results and has 8 number of attributes. It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/Immunotherapy+Dataset).
Anticancer Peptides Membranoid anticancer peptides (ACPs) are drawing increasing as potential cancer therapies due to their ability to prevent cellular resistance and overcome common hurdles such as chemotherapy side effects and cytotoxicity. The anticancer action of peptides (annotated for their one-letter amino acid code) on breast and lung cancer cell lines is described in this dataset. It is of a high standard (active, moderately active, experimental inactive, virtual inactive). It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/Anticancer+peptides).
Gene Expression Cancer RNA-Seq The samples are sorted by row. The RNA-Seq gene expression levels measured by the illumina HiSeq platform are the attributes of each sample. Number of attribute is 20531 for 801 sample, it is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/gene+expression+cancer+RNA-Seq).
Primary Tumour This is one of three domains provided by the Oncology Institutenthat has repeatedly appeared in the machine learning literature. For 339 instances, it contains 17 attributes. It is available on the UCI Web page (https://archive.ics.uci.edu/ml/datasets/primary+tumor).
Alzheimer Features This dataset is used to describe Alzheimer features. It consists of 347 instances with 10 features. This dataset is collected from kaggle website’s Web page (https://www.kaggle.com/datasets/brsdincer/alzheimer-features).
Eye Disorder This dataset discusses eye disorder. It is used to describe 101 instances for 16383 features. It is available on the kaggle website’s Web page (https://www.kaggle.com/datasets/prateek0x/eye-disorder-dataset).
5.3.2 Data pre-possessing
Some chemical data may require some preprocessing procedures (Fig. 7) which illustrates the main stages of the preparation process: (1) The information about proteins is transformed into a chemical representation; (2) descriptors are calculated; and (3) the chemical structure is converted into a mathematical form. The following are the phases.
-
1.
Protein information is transformed into an isomeric simplified molecular-input line entry system (SMILES) using the open Babel software [63]. The information about proteins is stored in a chemical format, called MOA, which must be transformed into isomeric SMILES using Babel software. Features are attributes with values used to create instances.
-
2.
E-Dragon is used to calculate the descriptors, and several chemical features are performed to implement several 2D and 3D data in the QSAR model and calculate descriptors using E-Dragon software. The descriptors are divided into three categories: rotary links, structural, and physicochemical links (weight and volume of molecule, distance between atoms, atom type, molecular account walking, electronegativity, atom distribution, aromatic, and thawed characteristics).
-
3.
The QSAR model expresses the mathematical relationship between chemical design and biological activity. Additionally, the features can be used to identify the instances. As shown in Fig. 8, QSAR is used to declare the major properties of chemical compounds. Moreover, structure-activity correlation analysis uses different ML algorithms to predict the similarity of chemicals in the presence of a disease. Compounds of complex molecules have many characteristics, such as topological factors [52].
5.3.3 Statistical results analysis
Table 3 presents the statistical criteria of the best value that was provided from the suggested algorithm mHGS compared with other algorithms for each dataset. Results in terms of mean, STD, best, worst, and CPU time values revealed the preference of the proposed algorithm in solving the FS problem compared to other algorithms. The best is the maximum value, the worst is the minimum, and STD is the smallest value which are depicted in Tables 4 and 5.
5.3.4 Convergence behaviour analysis
The convergence curves of the proposed mHGS have been compared to other algorithms. The comparison is presented in Fig. 9 for ten datasets. mHGS algorithm has a stable point for all datasets, suggesting that the proposed algorithm converges. Furthermore, mHGS has achieved the greatest average of the best solutions, the fastest for most datasets. The suggested mHGS algorithm is a promising optimization choice for solving FS problems and achieving high accuracy when compared to existing algorithms, as shown in Fig. 9.
5.3.5 Boxplot behaviour analysis
The boxplot is used to evaluate the performance of several dataset as a non-parametric method. However, in descriptive statistics, a boxplot is a way for graphically depicting groups of numerical data through their quartiles. Boxplots may also have lines extending vertically from the boxes, indicating variability outside the upper and lower quartets; thus, the terms “box-and-whisker plot” and “box-and-whisker diagram”. The maximum or minimum is the largest or the lowest data points achieved by the algorithm. Individual points can be plotted as outlines. The distance between the various parts of the box reflects the degree of spread and skewness in the data, as well as the contours of the data. In the experiments, the boxplots for mHGS-SVM over the ten datasets are presented in Fig. 10. The boxplots of the proposed mHGS algorithm are very narrow compared to other algorithm distributions for most datasets.
5.4 Discussion
Firstly, the proposed mHGS and other compared algorithms are assessed on the CEC’20 benchmark. After that, ten medical dataset are used to evaluate the proposed mHGS-SVM performance. For the CEC’20 benchmark, quantitative and qualitative metrics are used to assess mHGS performance. The proposed mHGS has achieved the highest value, but the IMODE algorithm achieved the lowest results for mean and STD statistical results, as shown in Table 3 and the best for convergence. The minimum convergence curve and boxplot as drawn in Figs. 4 and 5. Figure 3 shows the parameter space is used for 3D visualization of the CEC’20 functions to understand the differences and nature of each problem. The qualitative metrics are used to draw stronger conclusions regarding the algorithm performance for a real problem to confirm the high performance of the proposed mHGS algorithm as shown in Fig. 6.
For FS, the proposed mHGS-SVM maximizes accuracy and reduces the number of features. The mHGS-SVM achieved the best value for mean, STD, best, worst, and computational time as shown in Tables 4 and 5, over all the datasets. The evidence for this fact is supported by the convergence curves when it is possible to see that the mHGS-SVM over ten medical dataset, as illustrated in Fig. 9. The convergence curve is selected because it represents the relationship between the number of features and the fitness function. It indicates the best-performed algorithm from the comparison between different approaches. Boxplot analysis indicates that mHGS-SVM achieved the highest performance compared with other algorithms, as shown in Fig. 10.
According to the above analysis, the proposed mHGS-SVM approach has reached better results than other counterparts. The HGS is the second-ranked, whereas the IMODE is the last rank. For clear comparison and under the same parameter setting, the search agents number was set to 30 for all experiments with a different number of dimensions, according to different dataset dimensions.
5.5 Comparison with existing studies
This subsection will go over several algorithms for automating fuzzy modelling. Many algorithms can be used to automate fuzzy modelling. The comparison of MHs fuzzy logic algorithms is described in Table 6. When choosing meta-heuristic algorithms for fuzzy modelling, there are many elements to consider. Many elements play an important role to the algorithm’s ability, including the representation of fuzzy parameters, the interpretability of the fuzzy model produced by the algorithm. In addition to the algorithm’s parameters, such as population number and the specific parameters according to the algorithm itself, the process involved in the algorithm, and the algorithm’s processing speed. A comparison analysis of MHs to determine the strengths and drawbacks of these algorithms is shown in Table 7.
6 Conclusion and future directions
In this work, an approach to improve the original hunger games search algorithm (HGS) is offered by employing the fuzzy mutation method and linear reduction exploration to avoid local optimal and improve the balance between global and local search. The modified mHGS algorithm was tested on the standard benchmark CEC’20. mHGS was applied for a classification strategy and FS method in several biomedical datasets. The SVM method was used for classifying the data, and it had a high average accuracy rate of 98.060%. Furthermore, using the mHGS for FS significantly improved the SVM classification performance. The experimental results showed that the proposed method provides superior classification results than others. In the future studies, a multi-objective optimization algorithm will be implemented for HGS to solve the FS problem in high-dimensional biomedical datasets.
As future directions, the proposed mHGS algorithm can be utilized in the following future perspectives such as (1) solving other real-world and large-scale optimization problems, (2) solving different engineering and real-world problems with unknown search spaces, (3) tackle with different problems such as feature selection, parameter identification, and task scheduling, and (4) solving multi-objective problems can be investigated in the future studies.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
References
Heidari AA, Mirjalili S, Faris H, Aljarah I, Mafarja M, Chen H (2019) Harris hawks optimization: algorithm and applications. Future Gen Comput Syst 97:849–872
Wah YB, Ibrahim N, Hamid HA, Abdul-Rahman S, Fong S (2018) Feature selection methods: case of filter and wrapper approaches for maximising classification accuracy. Pertan J Sci Technol 26(1)
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28
Kennedy J, Eberhart RC (1997) A discrete binary version of the particle swarm algorithm. In: 1997 IEEE International conference on systems, man, and cybernetics. Computational cybernetics and simulation, vol 5, pp 4104–4108. IEEE
Gupta Y, Saini A (2017) A novel fuzzy-pso term weighting automatic query expansion approach using combined semantic filtering. Knowledge-Based Syst 136:97–120
Hancer Emrah, Xue Bing, Zhang Mengjie, Karaboga Dervis, Akay Bahriye (2018) Pareto front feature selection based on artificial bee colony optimization. Inf Sci 422:462–479
Rodrigues D, Pereira LAM, Almeida TNS, Papa JP, Souza AN, Ramos CCO, Yang X-S (2013) Bcs: A binary cuckoo search algorithm for feature selection. In 2013 IEEE International symposium on circuits and systems (ISCAS), pp 465–468. IEEE
Seyedali M, Mohammad MS, Andrew Lewis (2014) Grey wolf optimizer. Adv Eng Softw 69:46–61
Sallam KM, Elsayed SM, Chakrabortty RK, Ryan MJ (2020) Improved multi-operator differential evolution algorithm for solving unconstrained problems. In: 2020 IEEE congress on evolutionary computation (CEC), pp 1–8. IEEE
Rashedi E, Nezamabadi-Pour H, Saryazdi S (2009) Gsa: a gravitational search algorithm. Inf Sci 179(13):2232–2248
Mirjalili S, Lewis A (2016) The whale optimization algorithm. Adv Eng Softw 95:51–67
Li S, Chen H, Wang M, Ali AH, Seyedali M (2020) A new method for stochastic optimization. In: Future generation computer systems, slime mould algorithm
Huang C-L, Dun J-F (2008) A distributed pso-svm hybrid system with feature selection and parameter optimization. Appl Soft Comput 8(4):1381–1391
Yutao Y, Huiling C, Asghar HA, Gandomi Amir H (2021) Hunger games search: visions, conception, implementation, deep analysis, perspectives, and towards performance shifts. Expert Syst Appl 177:114864
Jun MB, Shuai L, Asghar HA (2022) Multi-strategy ensemble binary hunger games search for feature selection. Knowledge-Based Syst 248:108787
Onay FK, Berkan AS (2022) Chaotic hunger games search optimization algorithm for global optimization and engineering problems. Math Comput Simul 192:514–536
Sanjoy C, Kumar SA, Ratul C, Moumita S, Sukanta N (2022) Hswoa: an ensemble of hunger games search and whale optimization algorithm for global optimization. Int J Intell Syst 37(1):52–104
Xu B, Heidari AA, Kuang F, Zhang S, Chen H, Cai Z (2022) Quantum nelder-mead hunger games search for optimizing photovoltaic solar cells. Int J Energy Res
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Computational protein-ligand docking and virtual drug screening with the autodock suite. Nat Protoc 11(5):905
Mohammad HB, Khurshid A, Sudeep R, Jalaluddin MA, Mohd A, Mohammad HS, Saif K, Mohammad AK, Ivo P, Inho C (2016) Computer aided drug design: success and limitations. Curr Pharm Des 22(5):572–581
Akbar JA, Reza F, Saeed R, Jiuyong L, Reza S, Esmaeil E (2016) Drugminer: comparative analysis of machine learning algorithms for prediction of potential druggable proteins. Drug Discov Today 21(5):718–724
Shuguang Y, Stephen CHC, Slawomir F, Horst V (2016) Pymol and inkscape bridge the data and the data visualization. Structure 24(12):2041–2042
Gräßer F, Kallumadi S, Malberg H, Zaunseder S (2018) Aspect-based sentiment analysis of drug reviews applying cross-domain and cross-data learning. In: Proceedings of the 2018 international conference on digital health, pp 121–125
Jimena MM, Sol DJ, Ignacio P (2018) Biclustering as strategy for improving feature selection in consensus qsar modeling. Electr Notes Discret Math 69:117–124
Goh Garrett B, Hodas Nathan O, Abhinav V (2017) Deep learning for computational chemistry. J Comput Chem 38(16):1291–1307
Virginia Sabando María, Ignacio P, Soto Axel J (2019) Neural-based approaches to overcome feature selection and applicability domain in drug-related property prediction. Appl Soft Comput 85:105777
Goncalves MV, Thales K, Zarzana EG, Rufino OP, Maria HK (2019) Advances with support vector machines for novel drug discovery. Expert Opin Drug Discov 14(1):23–33
Grisoni Francesca, Consonni Viviana, Ballabio Davide (2019) Machine learning consensus to predict the binding to the androgen receptor within the compara project. J Chem Inf Model 59(5):1839–1848
Khozeimeh F, Alizadehsani R, Roshanzamir M, Khosravi A, Layegh P, Nahavandi S (2017) An expert system for selecting wart treatment method. Comput Biol Med 81:167–175
Francesca G, Neuhaus Claudia S, Miyabi H, Gisela G, Hiss Jan A, Masaaki K, Gisbert S (2019) De novo design of anticancer peptides by ensemble artificial neural networks. J Mol Model 25(5):1–10
Suriya M, Chandran V, Sumithra MG, Xiao-Zhi G, Elakkiya B, Akila M, Manoharan S (2021) Demnet: a deep learning model for early diagnosis of alzheimer diseases and dementia from mr images. IEEE Access 9:90319–90329
Hashim Fatma A, Houssein Essam H, Kashif H, Mabrouk Mai S, Walid A-A (2020) A modified henry gas solubility optimization for solving motif discovery problem. Neural Comput Appl 32(14):10759–10771
Hastie T, Tibshirani R, Friedman J, Franklin J (2005) The elements of statistical learning: data mining, inference and prediction. Math Intell 27(2):83–85
Wan Y, Wang M, Ye Z, Lai X (2016) A feature selection method based on modified binary coded ant colony optimization algorithm. Appl Soft Comput 49:248–258
Cai J, Luo J, Wang S, Yang S (2018) Feature selection in machine learning: a new perspective. Neurocomputing 300:70–79
Hussien AG, Hassanien AE, Houssein EH (2017) Swarming behaviour of salps algorithm for predicting chemical compound activities. In: 2017 eighth international conference on intelligent computing and information systems (ICICIS), pp 315–320. IEEE
Houssein EH, Hosney ME, Oliva D, Mohamed WM, Hassaballah M (2020) A novel hybrid harris hawks optimization and support vector machines for drug design and discovery. Comput Chem Eng 133:106656
Mohamed AE, Moemen Yasmine S, Ella HA, Shengwu X (2020) Toxicity risks evaluation of unknown fda biotransformed drugs based on a multi-objective feature selection approach. Appl Soft Comput 97:105509
Alaa T, Moemen Yasmine S, Ella HA (2016) A predictive model for toxicity effects assessment of biotransformed hepatic drugs using iterative sampling method. Sci Rep 6(1):1–13
Mohamed AE, Dalia Y (2021) Automatic selection of heavy-tailed distributions-based synergy henry gas solubility and harris hawk optimizer for feature selection: case study drug design and discovery. Artif Intell Rev 54:1–46
Kılıç F, Kaya Y, Yildirim S (2021) A novel multi population based particle swarm optimization for feature selection. Knowledge-Based Syst 219:106894
Nabil N, Houssein Essam H, Kashif H (2020) An efficient henry gas solubility optimization for feature selection. Expert Syst Appl 152:113364
Hashim Fatma A, Houssein Essam H, Mabrouk Mai S, Walid A-A, Seyedali M (2019) Henry gas solubility optimization: a novel physics-based algorithm. Futur Gener Comput Syst 101:646–667
Liu W, Yu X, Raj B, Yi L, Zou X, Li M (2015) Efficient autism spectrum disorder prediction with eye movement: a machine learning framework. In: 2015 International conference on affective computing and intelligent interaction (ACII), pp 649–655. IEEE
Fauzi IR, Rustam Z, Wibowo A (2021) Multiclass classification of leukemia cancer data using fuzzy support vector machine (fsvm) with feature selection using principal component analysis (pca). J Phys Conf Ser 1725:012012
Anter Ahmed M, Mumtaz A (2020) Feature selection strategy based on hybrid crow search optimization algorithm integrated with chaos theory and fuzzy c-means algorithm for medical diagnosis problems. Soft Comput 24(3):1565–1584
Lin L-C, Yeh Y-C, Chu T-Y (2014) Feature selection algorithm for ecg signals and its application on heartbeat case determining. Int J Fuzzy Syst 16(4)
Chen C, Li M, Sui J, Wei K, Pei Q (2016) A genetic algorithm-optimized fuzzy logic controller to avoid rear-end collisions. J Adv Transp 50(8):1735–1753
Yang B (2019) Dynamic risk identification safety model based on fuzzy support vector machine and immune optimization algorithm. Saf Sci 118:205–211
Ye H, Peiliang W, Zhu T, Xiao Z, Zhang X, Zheng Long, Zheng Rongwei, Sun Yangjie, Zhou Weilong, Qinlei Fu et al (2021) Diagnosing coronavirus disease 2019 (covid-19): efficient harris hawks-inspired fuzzy k-nearest neighbor prediction methods. IEEE Access 9:17787–17802
Hancer E, Xue B, Zhang M, Karaboga D, Akay B(2015) A multi-objective artificial bee colony approach to feature selection using fuzzy mutual information. In: 2015 IEEE congress on evolutionary computation (CEC), pp 2420–2427. IEEE
Luque RI, Aéngel G-NM (2017) Advantages of relative versus absolute data for the development of quantitative structure-activity relationship classification models. J Chem Inf Model 57(11):2776–2788
Kiralj R, Ferreira M (2009) Basic validation procedures for regression models in qsar and qspr studies: theory and application. J Braz Chem Soc 20(4):770–787
Honório Káthia M, De Lima EF, Quiles Marcos G, Romero Roseli AF, Molfetta Fábio A, Da Silva Albérico BF (2010) Artificial neural networks and the study of the psychoactivity of cannabinoid compounds. Chem Biol Drug Des 75(6):632–640
Moss GP, Shah AJ, Adams RG, Davey N, Wilkinson SC, Pugh WJ, Sun Y (2012) The application of discriminant analysis and machine learning methods as tools to identify and classify compounds with potential as transdermal enhancers. Eur J Pharm Sci 45(1–2):116–127
Rodriguez-Perez R, Vogt M, Bajorath J (2017) Support vector machine classification and regression prioritize different structural features for binary compound activity and potency value prediction. ACS Omega 2(10):6371–6379
Kar D, Ghosh M, Guha R, Sarkar R, Garcia-Hernandez L, Abraham A (2020) Fuzzy mutation embedded hybrids of gravitational search and particle swarm optimization methods for engineering design problems. Eng Appl Artif Intell 95:103847
Bouchet A, Montes S, Ballarin V, Díaz I (2020) Intuitionistic fuzzy set and fuzzy mathematical morphology applied to color leukocytes segmentation. Signal Image Video Process 14(3):557–564
Chowdhary CL, Acharjya DP (2016) A hybrid scheme for breast cancer detection using intuitionistic fuzzy rough set technique. Int J Healthc Inf Syst Inf 11(2):38–61
Senthil Kumar P (2020) Algorithms for solving the optimization problems using fuzzy and intuitionistic fuzzy set. Int J Syst Assur Eng Manag 11(1):189–222
Erdal K, Andriy S, Simon C, Robert J, Ahmadieh KM (2018) Type-2 fuzzy elliptic membership functions for modeling uncertainty. Eng Appl Artif Intell 70:170–183
Houssein Essam H, Emre Çelik, Mahdy Mohamed A, Ghoniem Rania M (2022) Self-adaptive equilibrium optimizer for solving global, combinatorial, engineering, and multi-objective problems. Expert Syst Appl 195:116552
Andersen JL, Flamm C, Merkle D, Stadler PF (2016) A software package for chemically inspired graph transformation. In: International conference on graph transformation, Springer, pp 73–88
Khan Asad U et al (2016) Descriptors and their selection methods in qsar analysis: paradigm for drug design. Drug Discover Today 21(8):1291–1302
Erdem D, Akgüngör Ali P (2016) Optimizing a fuzzy logic traffic signal controller via the differential evolution algorithm under different traffic scenarios. Simulation 92(11):1013–1023
Mahmoodabadi MJ, Danesh N (2018) Gravitational search algorithm-based fuzzy control for a nonlinear ball and beam system. J Contrand Decis 5(3):229–240
Kaur P, Kaur R (2020) Comparative analysis of classification techniques for diagnosis of diabetes. In: Advances in bioinformatics, multimedia, and electronics circuits and signals, Springer, pp 215–221
Ponce P, Soriano LA, Molina A, Garcia M (2018) Optimization of fuzzy logic controllers by particle swarm optimization to increase the lifetime in power electronic stages. In: Electric machines for smart grids applications-design, simulation and control
Lilla AD, Khan MA, Barendse P (2013) Comparison of differential evolution and genetic algorithm in the design of permanent magnet generators. In: 2013 IEEE International conference on industrial technology (ICIT), pp 266–271. IEEE
Derrac J, García S, Molina D, Herrera F (2011) A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol Comput 1(1):3–18
Woźniak M, Grana M, Corchado E (2014) A survey of multiple classifier systems as hybrid systems. Inf Fusion 16:3–17
Banaja M, Sasmita T (2016) A teaching learning based optimization technique for optimal location and size of dg in distribution network. J Electr Syst Inf Technol 3(1):33–44
Wahab MNA, Nefti-Meziani S, Atyabi A (2015) A comprehensive review of swarm optimization algorithms. PloS one 10(5):e0122827
Jahan SS, Sungur TS, Ibrahim E, Zakirhussain F (2018) Gravitational search algorithm (gsa) based pid controller design for two area multi-source power system load frequency control (lfc). Gazi Univ J Sci 31(1):139–153
Liu J, Xing Y, Li Y (2018) A gravitational search algorithm with adaptive mixed mutation for function optimization. Int J Perform Eng 14(4):681
Hu H, Cui X, Bai Y (2017) Two kinds of classifications based on improved gravitational search algorithm and particle swarm optimization algorithm. Adv Math Phys 2017
Shen D, Jiang T, Chen W, Shi Q, Gao S (2015) Improved chaotic gravitational search algorithms for global optimization. In: 2015 IEEE congress on evolutionary computation (CEC), pp 1220–1226. IEEE
Ning J, Zhang C, Sun P, Feng Y (2019) Comparative study of ant colony algorithms for multi-objective optimization. Information 10(1):11
Zeineb A, Adel G, Lazhar B-B, Mohamed H, Ahmed A-EN (2017) Review of optimization techniques applied for the integration of distributed generation from renewable energy sources. Renew Energy 113:266–280
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that there are no conflict of interest. Non-financial competing of interests.
Human or animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Houssein, E.H., Hosney, M.E., Mohamed, W.M. et al. Fuzzy-based hunger games search algorithm for global optimization and feature selection using medical data. Neural Comput & Applic 35, 5251–5275 (2023). https://doi.org/10.1007/s00521-022-07916-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07916-9