Abstract
Mitosis assessment of breast cancer has a strong prognostic importance and is visually evaluated by pathologists. The inter, and intra-observer variability of this assessment is high. In this paper, a two-stage deep learning approach, named MITNET, has been applied to automatically detect nucleus and classify mitoses in whole slide images (WSI) of breast cancer. Moreover, this paper introduces two new datasets. The first dataset is used to detect the nucleus in the WSIs, which contains 139,124 annotated nuclei in 1749 patches extracted from 115 WSIs of breast cancer tissue, and the second dataset consists of 4908 mitotic cells and 4908 non-mitotic cells image samples extracted from 214 WSIs which is used for mitosis classification. The created datasets are used to train the MITNET network, which consists of two deep learning architectures, called MITNET-det and MITNET-rec, respectively, to isolate nuclei cells and identify the mitoses in WSIs. In MITNET-det architecture, to extract features from nucleus images and fuse them, CSPDarknet and Path Aggregation Network (PANet) are used, respectively, and then, a detection strategy using You Look Only Once (scaled-YOLOv4) is employed to detect nucleus at three different scales. In the classification part, the detected isolated nucleus images are passed through proposed MITNET-rec deep learning architecture, to identify the mitosis in the WSIs. Various deep learning classifiers and the proposed classifier are trained with a publicly available mitosis datasets (MIDOG and ATYPIA) and then, validated over our created dataset. The results verify that deep learning-based classifiers trained on MIDOG and ATYPIA have difficulties to recognize mitosis on our dataset which shows that the created mitosis dataset has unique features and characteristics. Besides this, the proposed classifier outperforms the state-of-the-art classifiers significantly and achieves a \(68.7\%\) F1-score and \(49.0\%\) F1-score on the MIDOG and the created mitosis datasets, respectively. Moreover, the experimental results reveal that the overall proposed MITNET framework detects the nucleus in WSIs with high detection rates and recognizes the mitotic cells in WSI with high F1-score which leads to the improvement of the accuracy of pathologists’ decision.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer is the most common cancer in women other than the non-melanoma skin cancer. It is the fourth leading cause of cancer mortality. To predict the aggressiveness of the breast cancer, the histological grade is a significant prognostic factor [28]. It is obtained using Nottingham Grading System (NGS) by assessing three different tumor features which are tubule formation, nuclear pleomorphism, and mitotic rate [4]. According to the NGS, mitotic rate provides the strongest prognostic value of these three features [4, 8]. Hence, mitosis detection and recognition play an important role, not only in the accurate assessment of cancer diagnosis and grading but also for predicting the treatment effectiveness and survival [6]. Besides this, estimation of mitotic rate depends on the correct identification of mitotic nuclei in whole slide images (WSI) [4]. Such identification task is usually performed by pathologists visually, which is tremendously difficult, highly variable, intensive process, and time-consuming. Therefore, it is of great importance to develop an automated computer-aided system for mitosis classification.
To develop an automated computer-aided system, it is essential to detect nucleus first and then, recognize mitosis in WSIs accurately. However, this is a challenging task, since there are various complexities and difficulties which are (1) differences in staining among the samples from different laboratories, (2) variable features of slide scanners, (3) inter- and intra-observer variability, (4) divergent size and shape, and (5) different color features of nuclei and cells. In order to overcome all these issues, we propose a new, and efficient deep learning-based framework to solve the corresponding problem.
1.1 Related work
The detection and enumeration of mitotic cells require two stages: segmentation and classification [19]. Earlier works on segmentation mainly focused on three different approaches: (1) thresholding-based, (2) edge detection based, and (3) clustering-based segmentation [1]. One of the well-known and most used thresholding-based methods is Otsu’s method, and it is a global thresholding-based algorithm [32]. Region-based cell segmentation and boundary-based cell segmentation algorithms were suggested by Paul and Mukherjee. The authors also proposed a method of segmentation based on relative entropy between cells and their backgrounds with the help of opening and closing morphological operations [31]. Yang et al. [48] introduced a watershed algorithm using marker controlling to segment clustered cells efficiently. Other than morphological operations and entropy thresholding, contextual information from the objects in histopathological images was also used as a method to segment nuclei. Active contours and level set methods were among the methods for boundary-based segmentation. Three of the most common clustering-based algorithms used for segmentation in biomedical image analysis are k-means clustering, fuzzy c-means clustering, and Gaussian mixture models [1]. Most of these earlier works assume similarity in size and distinguishable color features of different cells. However, under certain conditions, nuclei enlarge and exhibit margination of chromatin while prominent nucleoli appear inside the nuclear boundary [21]. Lack of generalizing across a broad spectrum of tissue morphologies caused former methods to fail. Furthermore, handcrafted features might not be distinctive for the segmentation tasks due to staining variability in histopathological images. Deep learning models learn the related features in a hierarchical manner, which makes models more robust to textural and color changes in the histopathological images, compared to traditional methods.
Recently, many machine learning-based methods have been designed and proposed for mitotic cells detection within histopathological images [4, 25, 30, 36, 41]. Moreover, several mitoses detection challenges have been organized: MITOS12 [36], AMIDA13 [36], MITOS14, [36], and TUPAC16 [36] and these challenges have tackled gradually more complicated and complex scenarios and have significantly improved the mitosis detection research. Note that, even though the complexity increases in these datasets, there are several issues and limitations that are explained and discussed in the next section. The obtained results in these competitions, deep learning methods have shown great performance to achieve high detection rates of mitotic cells. Some of the latest algorithms have made use of deeper [30], wider [50] and cascades of networks [10]. Top performing methods in theses challenge are mostly based on convolutional neural networks (CNNs) [11, 20, 30]. According to the results in these challenges, the computer-aided systems based on deep CNNs are the top-performing approaches in mitosis detection and classification, and thus, have become the standard approach for automatic mitosis detection.
CNNs have proven to outperform traditional techniques such as conventional image processing methods including thresholding, region growing, graph cuts, k-means clustering, probabilistic models, active contours, and morphological operations in nuclei segmentation and mitosis detection [35, 37]. [11] are the first to utilize a CNN-based approach, CNN-based deep max-pooling, for mitosis detection in histopathological images of the breast. Having achieved an F-score of 0.78, they won the MITOS contest at ICPR 2012. Ciresan et al. [11] also won the AMIDA 2013 challenge with their Multi-Column Max-Pooling CNNs with an F-score of 0.61. The MITOS-ATYPIA-14 challenge had a different winner with a different approach called the Deep Cascade Network that is very similar to our proposed method. Chen et al. [9] firstly identified candidates for mitotic figure detection and then subsequent classification only focused on these candidates. They won the contest with an F score of 0.356. Xu et al. [47] integrated the SSAE (Stacked Sparse Autoencoder)-based framework for learning of high-level features associated with nuclei. An extensive survey of computer-aided methods for breast cancer diagnostic can be found in [12, 29].
Considering that a WSI consists of a vast number of high-power fields and mitoses are sparsely distributed, the pixel-wise classifier feature of CNN is time-consuming for mitosis detection when used alone. However, as Chen et al. [9] stated, a step of defining candidates or region of interest (ROI) can reduce the detection time and facilitate the use of the models in practice. Our method applied Scaled-Yolov4 [44] for the nuclei detection task. ROIs defined by Scaled-Yolov4, the nuclei of the cells, are used as inputs for the proposed deep learning-based classification framework for mitosis classification.
1.2 Contribution
This paper presents a new two-stage deep learning framework, named MITNET, to automatically classify mitosis in WSIs. The proposed framework consists of MITNET-det for nucleus detection and MITNET-rec for mitosis classification. In MITNET-det, the scaled-Yolov4 detection algorithm is used. In MITNET-rec, a new deep learning classifier is proposed. Moreover, to train and validate the proposed MITNET deep learning architectures, two new datasets which are nucleus dataset, and mitosis dataset are introduced and used (Fig. 1).
The main contributions and strengths of the paper are summarized as follows:
-
1.
Designing the MITNET framework based on deep learning network architectures which include MITNET-det and MITNET-rec. The MITNET-det uses scaled-Yolov4 [44] to detect the nucleus in WSIs of breast cancer. Moreover, to the best of our knowledge, this is the first successful attempt at applying scaled-YOLOv4 [44] to detect nuclei in WSIs. Note that, we have provided a web server link (http://212.156.134.202:4481/) where the users can upload images with the size of \(512 \times 512\) and run the proposed approach to detect nuclei and recognize mitosis cells,
-
2.
Developing a deep learning architecture (MITNET-rec) based on VGG-11 [34] convolutional neural network for mitosis recognition,
-
3.
Introducing two new datasets, which provide a large number of highly diverse, accurate and detailed annotations of nucleus in patches and extraction of image samples of mitotic cells. The first dataset is used to detect nucleus in the WSIs and consists of 139,124 annotated nucleus in 1749 patches from 115 WSIs of breast cancer, whereas the second one contains 4908 mitotic cells and 4908 non-mitotic cells which is used for mitoses classification. Moreover, the datasets are collected by three bio-engineers and two expert pathologists.
-
4.
Developing a time-saving and cost-effective AI-based computer-aided system for pathologists and doctors to effectively and accurately identify the mitoses in WSIs and thus, they can count the mitoses efficiently.
2 Mitosis recognition challenges and limitations of existing datasets
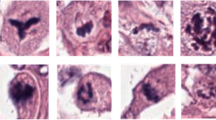
Visual assessment of mitotic cells using a light microscope is a time-consuming and difficult process which depends on the pathologists’ experience [24]. This is due to the fact that the pathologists generally must manually extract morphological features such as apoptotic cells, hyperchromatic structures, deformed nuclei and lymphocytes to distinguish the mitotic and non-mitotic cells in WSIs. However, due to existence of high complexity and artifacts, the detection and recognition accuracy is prone to error. The phases of normal mitosis include prophase, prometaphase, metaphase, anaphase, and telophase which have different morphological variations and, the mitosis observed in the glass slides can also be in a transition state between phases. Abnormalities during cell division can result in morphological appearances that differ from normal mitosis, called atypical mitosis (e.g., tripolar mitotic figures) [40]. Morphological features of mitosis and atypical mitosis need to be distinguished from mitosis-like figures (non-mitosis) during visual assessment by pathologists. However, it is often difficult to make this distinction by pathologists, so various AI-based computer aided systems have been developed to avoid subjectivity and difficulties in the mitosis detection and recognition process. In this respect, many competitions have been organized for automatic mitosis recognition and several mitosis datasets have been shared with researchers in recent years, e.g., the MICCAI-TUPAC 16 challenge, [43], ICPR MITOS-ATYPIA-2014 challenge [33], and MIDOG 2021 [23]. In addition to the publicly available mitosis datasets, there are several publicly available nuclei datasets. For instance, a nuclei dataset from the MoNuSeg (Multi-Organ Nuclei Segmentation) 2018 challenge [22] and the NuCLS dataset [2] are created for the research community to improve the performance of the AI-based systems. The major deficiency of publicly available nuclei datasets is the absence of images containing mitosis. Even though these datasets are crucial in the development of AI models for automatic nuclei and mitosis detection and recognition, they have some limitations such as (1) variability of images caused by the specimen preparation, (2) varying staining process, (3) WSI quality, (4) lack of available complete (both nuclei and mitosis) datasets and (5) limited number of samples in available datasets. Moreover, these datasets have been re-examined by an expert pathologist who confirmed that these datasets include false positive mitoses samples (see Fig. 2). For instance, as shown in Fig. 2, apoptotic figures, non-mitotic nuclei, blurry and pinkish figures are abundantly available in publicly available datasets. Some factors such as artifacts due to staining techniques and digitization by different scanners during the preparation of WSIs may cause incorrect mitosis annotations. Therefore, it is essential to create a new dataset from different WSIs to further develop automated AI-based mitosis detection and recognition models.
3 In-house dataset collection and description
To avoid the aforementioned limitations and issues of the existing datasets as well as increase the accuracy of the automated AI-based models, we have created a new dataset which consists of (1) annotated nuclei in patches of WSIs, and (2) extracted mitosis and non-mitosis image samples from WSIs. The in-house dataset contains different sizes, shapes and colors of annotated nucleus on different patches extracted from WSIs of breast cancer tissue (see Fig. 3). In addition, since the publicly available mitosis datasets contain a confined variety of mitotic image samples in terms of color, size, shape, and variation, a new mitosis dataset is generated by collecting mitotic and non-mitotic image samples from WSIs (see Figs. 1 and 4). It is important to note that to collect the in-house dataset, the WSIs are scanned by 3DHISTECH scanner at 20\(\times\) magnification which minimizes the confounding effect of variance in staining methods and the experience of operating personnel. On the other hand, MIDOG dataset has been extracted from WSIs scanned by Hamamatsu XR nanozoomer 2.0, Hamamatsu S360 and Aperio ScanScope CS2 scanners, and MITOS-ATYPIA-14 Aperio dataset has been extracted from WSIs scanned by Aperio Scanscope XT scanner at 40\(\times\) magnification.
The annotations of nuclei in the patches and extraction of mitosis image samples have been carried out manually by three bioengineers and two expert pathologists in the field of breast pathology. The in-house dataset consists of 139,124 annotated nuclei in 1749 different patches with the size of \(512 \times 512\) pixels which are extracted from 115 different WSIs with the size of \(87,780 \times 109,494\) pixels. Note that, each WSI has been obtained from different patient. More specifically, 115 WSIs have been obtained from 115 different patients. The in-house mitosis classification dataset is created as a unique dataset that contains 4908 mitotic and 4908 non-mitotic image samples with the size of \(50 \times 50\). We collected these image samples from 214 WSIs which have been obtained from 214 different patients. Due to the variations in quality of images in the collected dataset, deep learning models cannot be trained efficiently and effectively. To tackle this problem, a color normalization technique [42] is used as a data pre-processing step to improve the color appearance and contrast of low-quality histology patches. Afterwards, five different augmentation techniques including horizontal flip, vertical flip, mosaic, scaling, and HSV (Hue, Saturation, Value) are applied to the created nuclei and mitosis datasets to increase the diversity and amount of the training data for improving the performance of MITNET framework. After the augmentations are applied, the nuclei and mitosis datasets have become five times larger. In the testing part, the proposed two-stage deep learning framework was performed on five different WSIs which include 1021 annotated mitotic nuclei and 1021 annotated non-mitotic nuclei.
4 MITNET: proposed methodology for nuclei detection and mitosis recognition
The main purpose of this work is to develop an automated computer-aided system using deep learning techniques for mitosis classification in the WSI of breast cancer tissue (users can upload images with the size of \(512 \times 512\) and run the proposed approach on the given web server link http://212.156.134.202:4481/). To achieve it, the aforementioned two datasets have been first created and then, used to train and validate the proposed deep learning framework, named MITNET, to detect nucleus and classify mitosis accurately in WSIs. In the proposed approach, the nuclei in the histopathological images of the breast tissue are detected using the MITNET detection approach (MITNET-det) based on scaled-Yolov4 architecture, whereas, in the MITNET recognition (MITNET-rec), a new deep learning architecture is designed and applied to determine whether the detected nucleus is classified as mitosis or non-mitosis. In general, flowchart of the proposed mitosis recognition methodology is illustrated in Fig. 5.
4.1 MITNET-det: nuclei detection in whole slide images of breast cancer
In the proposed approach, detection of nuclei in WSIs is a vital prerequisite task for automated mitosis classification. In this work, to detect nuclei in images, an object detection algorithm is used. Generally, the-state-of-the-art object detection algorithms can be classified into three categories: (1) conventional computer vision based, (2) two-stage deep learning-based, and (3) single-stage deep learning-based algorithms. Conventional object-detection methods are mainly based on sliding window search and handcrafted features. This approach results in low detection accuracy and is computationally expensive. In the two-step-approach, a region proposal architecture is firstly employed to predict candidate object bounding boxes. After that, a CNN model is used to extract features from each predicted candidate box and classify them using regression technique. Despite the two-step approach shows high detection accuracy, it is computationally expensive. The last object detector category applies a single deep CNN model with regression technique to simultaneously detect and classify objects in images. Moreover, this approach provides decent accuracy and has the lowest computational cost when compared to the other object detection categories. With regard to the existing one-step algorithms, You Only Look Once (YOLO) series algorithms have been used in many different applications and achieved higher mean average precision (mAP) than the other one-step deep learning-based detection algorithms.
Therefore, in this paper to automatically detect nuclei in WSIs, a YOLO-based deep learning framework, named MITNET-det, is used. There are various versions of YOLO algorithms. Among all the YOLO methods, scaled Yolov4 [5] is one of the most efficient object detection methods which was developed recently. Chien et al. [44] proposed a network scaling approach that modifies not only the depth, width, resolution, but also structure of the network. The scaled YOLOv4 is composed of three architectures: backbone, neck, and head. The backbone architecture aims at extracting features from input image. To achieve this, the backbone structure of scaled YOLOv4 uses a convolutional neural network that combines Darknet53 with Cross Stage Partial Network (CSPNet) to partition the feature map of each layer into two parts and then fused them through a cross-stage hierarchy. The CSPNet allows for more gradient flow through the network which reduces the amount of computation and achieves a trade-off between speed and accuracy [45]. The neck architecture concatenates the feature maps from different layers of the CSPNet-Darknet53 backbone network and delivers them as inputs to the head network. To this end, the neck combines CSPNet-Path Aggregation Network (PAN) with CSPNet-Spatial Pyramid Pooling (SPP) modules. The CSPNet-SPP network utilizes four different maximum pooling layers to generate feature maps with different scales, whereas the CSPNet-PAN uses a shortcut connection to fuse these feature maps. Finally, the generated three scales feature maps are fed into the head layer which is similar to YOLOv3 to predict bounding box, class, and confidence score. The main principle of the scaled YOLOv4 is to analyze quantitative costs of various CNN models while changing image sizes, number of layers, and number of channels. One other improvement is to increase the number of stages of the network model to achieve a better accuracy for predicting larger objects in images. As a result, it is fast and provides better object detection accuracy. The further details of the scaled YOLOv4 algorithm can be found in [44].
Before training the algorithm, five different augmentation techniques are employed to increase the diversity and the size of the nuclei dataset. Thus, the performance of the trained detection algorithm has been further improved. Furthermore, the scaled-YOLOv4 [44] has been applied to detect objects in many application areas including remote sensing [49], natural scene images [26], and many others [7, 38, 46]. To the best of our knowledge, this study is the first successful attempt at applying scaled-YOLOv4 to detection of nuclei in WSIs.
4.2 MITNET-rec: mitosis recognition network architecture
In the MITNET-rec, a mitosis classifier based on the CNN is developed and applied to classify the detected nucleus as mitosis or not-mitosis. Besides of the MITNET-rec, five different deep learning methods including Resnet50 [16], Resnet101 [16], Densenet161 [18], efficientnetB0 [39], and efficientnetB3 [17] are used for comparison purposes. Note that, all the classifiers used in this work are previously trained or pre-trained with ImageNet [13] consisting of over a million images for a wide range of objects. Further details of the proposed classifier and the compared deep learning classifiers are explained below:
-
Proposed CNN Architecture (MITNET-rec): This method is designed and developed based on the concept of VGG-11 CNN model [34]. The proposed mitosis classifier consists of a number of layers including convolutional, max-pooling, fully connected (FC) layers, and softmax layer. The training is performed with the Stochastic Gradient Descent optimization algorithm with mini-batches of 4 instances. The proposed CNN classification architecture consists of eight convolutional layers, four FC layers and a softmax layer. Each convolutional layer uses \(3 \times 3\) filters. The convolution stride is fixed to 1, and Max-pooling is performed with a \(2 \times 2\) window. Figure 6 shows each step of the proposed architecture. Of the FC layers, the first three layers consist of 4096, 2048, and 1024 neurons, respectively. The last layer performed two-way classification to classify the detected nuclei as mitotic nuclei or non-mitotic nuclei. Moreover, Rectified Linear Unit (ReLU) is applied as an activation function in all layers of the proposed deep learning model. Besides these, adaptive average pooling operation is used in the proposed model and the input images are with the size of \(50 \times 50\) pixels.
-
Resnet50 and Resnet101 [16]: Researchers tend to increase the depth of CNNs in order to improve the accuracy. However, it has been observed that the performance degrades. This is because the information about the gradient passing through many layers can vanish. He et al. [16] introduced a deep residual learning framework which explicitly let few stacked layers fit a residual mapping. They realized such an algorithm by using skip connections. The skip connections simply perform identity mapping, and their outputs are added to the outputs of the stacked layers. ResNet-34 was the first ResNet architecture inspired by VGG-19 including skip connections. The creators of ResNet-34 modified the building block due to the concerns on training time. They obtained ResNet-50 by using a stack of three layers instead of the earlier two for skip connections. ResNet-101 was constructed by using more 3-layer blocks.
-
Densenet161 [18]: As in ResNet, the key characteristic of the solution to tackle the vanishing gradient problem for deeper CNNs to create short paths from early layers to later layers. Huang et al. [18] distilled this insight into a simple connectivity pattern. They connected all layers (with matching feature-map sizes) directly with each other to ensure maximum information flow between layers in the network. Densenet concatenates the output feature maps of a layer with the incoming feature maps instead of summation like in ResNet.
-
EfficientnetB0 and EfficientnetB3 [17, 39]: CNNs are scaled up for better accuracy by increasing the layers if more resources are available. Increasing the width and depth of CNN was usually done manually and arbitrarily without considering a systematical approach. Tan and Le [39] proposed a new scaling method that uniformly scales all dimensions of depth/width/resolution using a simple yet highly effective compound coefficient. They scaled up the mobile inverted bottleneck convolution (MBConv) as a baseline to obtain a family of deep learning models, called EfficientNets [17]. All EfficientNet models are scaled from baseline EfficientNet-B0 using a different compound coefficient.
5 Experimental results
5.1 Experimental setup
5.1.1 Datasets
In order to understand, evaluate and analyze the performance of the proposed classification framework over the state-of-the-art methods, various datasets such as MIDOG [27], ATYPIA [3], and the in-house dataset have been used for mitosis classification. Note that, the publicly available MIDOG and ATYPIA datasets do not contain nuclei datasets.
The in-house dataset consists of 139,124 annotated nuclei in 1749 patches with the size of \(512 \times 512\) pixels extracted from 115 WSIs with the size of \(87,780 \times 109,494\) pixels. Note that, the 115 WSIs obtained from 115 different patients have been first randomly split into \(90\%\) for training and \(10\%\) for validation. After that, the patches with the size of \(512 \times 512\) have been extracted from the WSIs. Moreover, five different augmentation techniques are applied to the training annotated set before training the MITNET-det.
For the mitosis classification task, the in-house mitosis dataset and two publicly available datasets MIDOG and ATYPIA are used. The in-house mitosis classification dataset has 4908 mitotic and 4908 non-mitotic image samples with the size of \(50 \times 50\), whereas the MIDOG dataset consists of 1721 mitotic and 2714 non-mitotic samples with the size of \(50 \times 50\) collected from 150 patches, and ATYPIA dataset includes 749 mitotic image samples with the size of \(50 \times 50\) extracted from 1200 patches of WSIs. To generate the in-house mitosis dataset, 214 WSIs which are obtained from 214 different patients have been used. The WSIs have been first randomly split into \(90\%\) for training and \(10\%\) for validation. After that, the in-house mitosis dataset has been generated from the training and validation sets. For the MIDOG and ATYPIA datasets, we used \(90\%\) of the image samples for training and \(10\%\) for validation. Moreover, five augmentation techniques are applied to the mitosis image samples to increase the diversity and the size of the dataset before training the proposed classifier MITNET-rec and state-of-the-art classifiers. In the testing part, to understand and analyze the performance of the proposed two-stage deep learning approach and achieve the results, five different WSIs scanned by a 3DHISTECH scanner at 20\(\times\) magnification have been used and 1021 mitotic nuclei and are annotated on these WSIs.
5.1.2 Hyperparameters
The hyperparameter values have been obtained empirically based on the performance on the validation set in both MITNET-det and MITNET-rec. Besides this, to avoid overfitting, two different strategies have been considered in this work. In the first strategy, we focused on training data. Augmentation techniques have been used to increase the variety of image samples in the collected dataset. In the second strategy, we used early stopping and weight decay. Note that, weight decay adds penalty term to the cost function in the proposed method to shrink the weights during back-propagation. The value of the weight decay is 0.0005.
In the MITNET-dec, the parameter values such as initial learning rate, batch size and epoch are set to 0.01, 16, and 150, respectively. Moreover, to optimize learning rate and model weights learning rate scheduler (LambdaLR) and Stochastic Gradient Descent (SGD) are applied. Training is stopped when there is no change in mAP after 7 consecutive epochs. Also, k-means clustering is used to get the optimal dimensions of anchor boxes in the scaled-YOLOv4. For classifiers, the learning rate, batch size and epoch are set to 0.00001, 4, and 100, respectively. The SGD is used as optimizer to obtain the minimum value of the cost function. The models are trained and tested on a computer with a single NVIDIA GTX 3090 GPU, an Intel i9-8950HK CPU, and 16 GB RAM.
5.2 Performance of nuclei detection method
The first experiment focuses on understanding and analyzing the performance of the scaled-Yolov4 (MITNET-det) for nuclei detection by observing the precision and recall values. To achieve the results, the MITNET-det model was trained on the augmented annotated nuclei dataset and validated on the validation nuclei dataset. The MITNET-det model was evaluated by the mean Average Precision (mAP). If a predicted bounding box has at least 0.5 Intersection over Union (IoU) with the ground truth bounding box, the predicted bounding is considered to be a true positive (TP). An undetected ground truth bounding box is considered false negative (FN), while a predicted bounding box that does not match its ground truth bounding box with at least 0.5 IoU is considered to be a false positive (FP). TP, FN, and FP are used to calculate precision and recall values. Figure 7 demonstrates the precision-recall curve. Moreover, the results show that the MITNET-det model provides promising performance for nuclei detection with an mAP of 0.88 at IoU=0.5.
5.3 Comparison of deep learning-based classifiers on existing mitosis datasets
In the second experiment, a preliminary assessment and evaluation are conducted on publicly available MIDOG dataset. To achieve the results, six different deep learning-based image classifiers which are proposed MITNET-rec, densenet161, resnet50, resnet 101, efficientnet-B0 and efficientnet-B3 are applied to classify mitosis and not-mitosis images. More precisely, the compared deep learning-based classifiers are trained and tested on MIDOG mitosis dataset. The obtained precision, recall, and F1-score results are illustrated in Fig. 8. According to the results, the proposed MITNET-rec architecture provides the best performance with precision \(58.6\%\), recall \(82.9\%\), and F1-score \(68.7\%\). The second best performance is obtained using densenet161 with precision \(48.6\%\), recall \(77.7\%\), and F1-score 59.8, whereas the resnet101 provides the third best performance with precision \(36.8\%\), recall \(73.4\%\) and F1-score \(49.1\%\). The lowest scores belong to the resnet50, efficientnet-B0 and efficientnet-B3 classifiers. Consequently, the results show that the proposed MITNET-rec classifier achieves the greatest mitosis classification rate over the publicly available MIDOG dataset.
5.4 Performance of different deep learning classifiers on various mitosis datasets
The purpose of this experiment is to understand and analyze the diversities and similarities of various mitosis datasets. To achieve it, six different deep learning methods are used to classify mitosis and not-mitosis in five different WSIs. Also, three separate cases are considered for evaluating and assessing the deep learning classifiers; (case 1) trained on MIDOG mitosis dataset and tested on our in-house mitosis dataset, (case 2) trained on ATYPIA mitosis dataset and tested on our in-house mitosis dataset, (case 3) trained and tested on our in-house mitosis dataset. The obtained overall scores on five WSIs and average scores of the classifiers are tabulated in Tables 1, 2, and 3. According to the results, the lowest precision, recall, and F1-scores are obtained by classifiers in case 1 and case 2 which indicate that there are numerous varieties and diversities between the mitosis image samples in the publicly available datasets (MIDOG and ATYPIA) and in-house dataset. More precisely, these low scores reveal that the mitosis samples in the in-house dataset consists of unique features and characteristics. Moreover, they are more challenging to classify by using the deep learning classifiers because the classifiers generated by the publicly available datasets cannot accurately classify the mitosis samples in the in-house dataset. As tabulated in Table 3, the greatest average precision, recall, and F1-scores are obtained using the proposed MITNET-rec architecture with the in-house dataset which are \(75.4\%\), \(38.1\%\), and \(49\%\), respectively. However, the efficientnet-B0 yields the poorest performance with average precision, recall and F1-scores of \(36.3\%\), \(30.1\%\), and \(33.4\%\), respectively. Consequently, the obtained scores demonstrate that the deep learning classifiers trained on the existing datasets and tested on the created dataset cannot perform well.
5.5 Performance of the proposed MITNET framework on whole slide images
The purpose of this experiment is to qualitatively analyze the efficiency and robustness of the proposed MITNET framework. Besides this, the MITNET-rec is qualitatively compared with the densenet161 classifier which provides the second-best mitosis classification results as shown in Fig. 8 and Table 3. To achieve it, the augmented and annotated nuclei dataset has been used to train the MITNET-det, and the augmented mitosis dataset is used to train the MITNET-rec and densenet161 classifiers, respectively. In testing, the WSIs with the size of \(87,780 \times 109,494\) pixels are used as input for the proposed framework. In the system, the smaller patches with the size of \(512 \times 512\) are first automatically extracted from the WSIs. After that, the MITNET deep learning framework is applied to all patches. The qualitative results of the proposed MITNET framework are shown in Fig. 9. The results demonstrate that the MITNET-det detects the nuclei in the patches and localize them with boundaries accurately. After that, the detected and localized nuclei images are center cropped to \(50 \times 50\) which are used as input for the classifiers. The qualitative results indicate that the proposed MITNET-rec classifier obtains the mitotic cells accurately. In the second qualitative results as illustrated in Fig. 10, the detected nuclei in six different patches are used as input for MITNET-rec and densenet161 classifiers. Note that, these patches are assessed by three bioengineers and two expert pathologists and the ground truths are shown in Fig. 10. According to the bioengineers and pathologists, two patches shown in the first two rows of Fig. 10 do not contain any mitotic cells. However, the rest of the patches illustrated in the last four rows of Fig. 10 consists of a single mitosis. For the first two image patches, the densenet161 classifies a non-mitotic cell as a mitotic cell, whereas the proposed MITNET-rec does not find any mitotic cells. The image patch in the third row of the figure consists of a single mitosis, and both classifiers successfully classify the mitotic cell. On the other hand, for the rest of the patches, the proposed MITNET-rec classifies and localizes the mitotic cells correctly, but the densenet161 fails and cannot classify any mitotic cell in these patches. Consequently, even though densenet161 classifies some mitotic cells correctly, it has a high false positive rate which increases complexity for doctors and pathologists.
6 Conclusions
In this paper, a new dataset and a two-stage deep learning framework named MITNET are introduced. The dataset is manually collected from different wholes slide images (WSIs) by three bioengineers and two pathologists. The dataset consists of: (1) annotated nuclei in patches, (2) extracted mitosis and not-mitosis image samples. In addition to generated dataset, MITNET is presented for nuclei detection (MITNET-det) and mitosis classification (MITNET-rec) in WSIs. The MITNET-det uses scaled-Yolov4 which is trained on the annotated nucleus dataset. The results demonstrate that the model provides high nuclei detection rate. In the MITNET-rec, a new deep learning-based classifier based on VGG-11 is proposed for mitosis recognition in WSIs. Besides this, in this paper, six different pre-trained deep learning-based classifiers trained on various mitosis datasets (MIDOG) and tested on in-house dataset are evaluated and investigated. The obtained results reveal that the classifiers provide poor recognition performance. As a result, the mitosis samples in the in-house dataset contain unique characteristics and features as compared to the publicly available mitosis datasets. Furthermore, the proposed MITNET framework is employed on WSIs, and the results demonstrate that the proposed framework achieves high detection and classification rates. Consequently, the proposed MITNET framework provides a practical and convenient computer-aided system for helping doctors and pathologists to make their decisions more accurate and efficient.
Data availability
Accessing to the in-house nuclei detection and mitosis classification dataset are available upon reasonable request to academic investigators without relevant conflicts of interest for non-commercial use who agree not to distribute the data. The source codes used in this work are publicly available and can be downloaded from the given link (https://github.com/virasoft/MITNET/). Moreover, we have provided a web server link (http://212.156.134.202:4481/) where the users can upload images with the size of \(512 \times 512\) and run the proposed approach to detect nuclei and recognize mitosis cells. Access requests and queries about the dataset and code can be made to sercan.cayir@virasoft.com.tr.
References
Albayrak A, Bilgin G (2016) Mitosis detection using convolutional neural network based features. In: Proceedings of the 2016 IEEE 17th international symposium on computational intelligence and informatics (CINTI), pp 335–340
Amgad M, Atteya LA, Hussein H, et al (2021), NuCLS: a scalable crowdsourcing, deep learning approach and dataset for nucleus classification, localization and segmentation. arXiv preprint arXiv:2102.09099, pp 1–45
ATYPIA dataset. https://mitos-atypia-14.grand-challenge.org/Donwload/
Balkenhol MCA, Tellez D, Vreuls W et al (2019) Deep learning assisted mitotic counting for breast cancer. Lab Invest 99:1596–1606
Bochkovskiy A, Wang C-Y, Liao H-YM (2020) YOLOv4: optimal speed and accuracy of object detection. CoRR abs/2004.10934
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Cai Y et al (2021) YOLOv4-5D: an effective and efficient object detector for autonomous driving. IEEE Trans Instrum Meas 70:1–13
Chang JM, McCullough AE, Dueck AC, Kosiorek MS et al (2015) Back to basics: traditional Nottingham grade mitotic counts alone are significant in predicting survival in invasive breast carcinoma. Ann Surg Oncol 22(3):S509–S515
Chen H, Dou Q, Wang X, Qin J, Heng P-A (2016) Proceedings of the thirtieth AAAI conference on artificial intelligence mitosis detection in breast cancer histology images via deep cascaded networks. AAAI Press, pp 1160–1166
Chen H, et al (2016) Mitosis detection in breast cancer histology images via DeepCascaded networks. In: AAAI, pp 1160–1166
Cireşan D et al (2013) Medical image computing and computer-assisted intervention: MICCAI 2013 mitosis detection in breast cancer histology images with deep neural networks. Springer, Berlin Heidelberg, pp 411–418
Debelee TG et al (2020) Survey of deep learning in breast cancer image analysis. Evolv Syst 11(1):143–163
Deng J et al (2009) ImageNet: a large-scale hierarchical image database. In: IEEE conference computing vision pattern recognition (CVPR), pp 248–255. https://doi.org/10.1109/cvpr.2009.5206848
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5):403–410
Graham S et al (2021) Lizard: a large-scale dataset for colonic nuclear instance segmentation and classification. IEEE/CVF Int Conf Comput Vis Workshops (ICCVW) 2021:684–693
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE Conf Comput Vis Pattern Recognit (CVPR), pp 770–778
Howard AG, Zhu M, Chen B, et al (2017) MobileNets: efficient convolutional neural networks for mobile vision applications. ArXiv preprint arXiv:1704.04861, pp 1–9
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition (CVPR), pp 2261–2269
Jimênez G, Racoceanu D (2019) Application to mitosis analysis in breast cancer grading frontiers in bioengineering and biotechnology. Deep Learn Semant Segment Classif Comput Pathol 7(145):1–12
Khan AM, Eldaly H, Rajpoot NM (2013) A gamma-gaussian mixture model for detection of mitotic cells in breast cancer histopathology images. J Pathol Inform 4(11):1–6
Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imag 36(7):1550–1560
Kumar N, Verma R, Anand DZ et al (2019) A multi-organ nucleus segmentation challenge. IEEE Trans Med Imag 39(5):1380–1391
Lafarge MW, Koelzer VH (2021) Rotation invariance and extensive data augmentation: a strategy for the mitosis domain generalization (MIDOG) challenge. Biomedical image registration, domain generalisation and out-of-distribution analysis. Lecture Notes in Computer Science, pp 62–67
Lashen A, Ibrahim A, Katayama A, Ball G, Mihai R, Toss M, Rakha E (2021) Visual assessment of mitotic figures in breast cancer: a comparative study between light microscopy and whole slide images. Histopathology 79(6):913–925
Mahmood T, Arsalan M, Owais M, Lee MB, Park KR (2020) Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and deep CNNs. J Clin Med 9(3):1–25
Mennatullah H, Heba K, Hossam F (2021) Image colorization using Scaled-YOLOv4 detector. Int J Inform Sci Comput 21(3):107–118
MIDOG dataset. https://imig.science/midog/the-dataset/
Murugesan B, et al (2019) Deep detection and classification of mitotic figures. In: Proceedings of the SPIE conference on medical imaging—digital pathology, pp 177 – 182
Oza P, Sharma P, Patel S et al (2022) Deep convolutional neural networks for computer-aided breast cancer diagnostic: a survey. Neural Comput Appl 34:1815–1836
Paeng K, Hwang S, Park S, Kim M (2017) A unified framework for tumor proliferation score prediction in breast histopathology. Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, New York, pp 231–239
Paul A, Mukherjee DP (2015) Mitosis detection for invasive breast cancer grading in histopathological images. IEEE Trans Image Process 24(11):4041–4054
Qu Z, Zhang L (2010) Research on image segmentation based on the improved Otsu algorithm. Second Int Confer Intell Hum Mach Syst Cybern 2:228–231
Racoceanu D, Calvo J, Attieh E, Le Naour G, Gloaguen A (2014) Detection of mitosis and evaluation of nuclear atypia score in breast cancer histological images. In: International conference on pattern recognition (ICPR), pp 1–8
Simonyan K, Andrew Z (2015) Very deep convolutional networks for large-scale image recognition. In: Proceedings of the 3rd international conference on learning representations (ICLR2015), pp 1–14
Sirinukunwattana K, Raza SEA, Tsang Y, Snead DRJ, Cree IA, Rajpoot NM (2016) Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imag 35(5):1196–1206
Sohail A, Khan A, Wahab N et al (2021) A multi-phase deep CNN based mitosis detection framework for breast cancer histopathological images. Sci Rep 11(6215):1–18
Song Y, Zhang L, Chen S, Ni D, Lei B, Wang T (2015) Accurate segmentation of cervical cytoplasm and nuclei based on multiscale convolutional network and graph partitioning. IEEE Trans Biomed Eng 62(10):2421–2433
Sun H, Xing G (2021) A YOLOv4-based vehicle detection method from UAV Videos. In: Proceedings of the 2021 33rd Chinese control and decision conference (CCDC), pp 3082–3087
Tan M, Le Q (2019) EfficientNet: rethinking model scaling for convolutional neural networks. Int Conf Mach Learn 6105–6114
Taryn AD, Frances MM, Christof AB, Richard L et al (2021) Mitotic figures-normal, atypical, and imposters: a guide to identification. Vet Pathol 58(2):243–257
Tellez D, Balkenhol M et al (2018) Whole-slide mitosis detection in HE breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans Med Imag 37(9):2126–2136
Vahadane A et al (2016) Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imag 35(8):1962–1971
Veta M, Heng YJ, Stathonikos N et al (2019) Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge. Med Image Anal 54:111–121
Wang C-Y, Bochkovskiy A, Mark Liao H-Y (2020) Scaled-YOLOv4: scaling cross stage partial network. CoRR, abs/2011.08036
Wang C-Y, Mark Liao H-Y, Wu Y-H, et al (2019) CSPNet: a new backbone that can enhance learning capability of CNN. In: IEEE/CVF conference on computer vision and pattern recognition workshops (CVPRW), pp 1571–1580
Wang C-Y, Bochkovskiy A, Liao H-YM (2021) Scaled-YOLOv4: scaling cross stage partial network. IEEE/CVF Conf Comput Vis Pattern Recognit (CVPR) 2021:13024–13033
Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, Madabhushi A (2015) Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans Med Imag 35(1):119–130
Yang X, Li H, Zhou X (2006) Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and Kalman filter in time-lapse microscopy. IEEE Trans Circuits Syst I Regul Pap 53(11):2405–2414
Zakria Z, Deng J, Kumar R, Khokhar MS, Cai J, Kumar J (2022) Multiscale and direction target detecting in remote sensing images via modified YOLO-v4. IEEE J Select Top Appl Earth Observ Remote Sens 15:1039–1048
Zerhouni E, Lanyi D, Viana M, Gabrani M (2017) Wide residual networks for mitosis detection. In: IEEE international symposium on biomedical imaging, pp 924–928
Acknowledgements
This project is funded by the Acibadem Hospital in Turkey.
Funding
Open access funding provided by Blekinge Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Çayır, S., Solmaz, G., Kusetogullari, H. et al. MITNET: a novel dataset and a two-stage deep learning approach for mitosis recognition in whole slide images of breast cancer tissue. Neural Comput & Applic 34, 17837–17851 (2022). https://doi.org/10.1007/s00521-022-07441-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07441-9