Abstract
Non-motor manifestations of Parkinson’s disease (PD) appear early and have a significant impact on the quality of life of patients, but few studies have evaluated their predictive potential with machine learning algorithms. We evaluated 9 algorithms for discriminating PD patients from controls using a wide collection of non-motor clinical PD features from two databases: Biocruces (96 subjects) and PPMI (687 subjects). In addition, we evaluated whether the combination of both databases could improve the individual results. For each database 2 versions with different granularity were created and a feature selection process was performed. We observed that most of the algorithms were able to detect PD patients with high accuracy (>80%). Support Vector Machine and Multi-Layer Perceptron obtained the best performance, with an accuracy of 86.3% and 84.7%, respectively. Likewise, feature selection led to a significant reduction in the number of variables and to better performance. Besides, the enrichment of Biocruces database with data from PPMI moderately benefited the performance of the classification algorithms, especially the recall and to a lesser extent the accuracy, while the precision worsened slightly. The use of interpretable rules obtained by the RIPPER algorithm showed that simply using two variables (autonomic manifestations and olfactory dysfunction), it was possible to achieve an accuracy of 84.4%. Our study demonstrates that the analysis of non-motor parameters of PD through machine learning techniques can detect PD patients with high accuracy and recall, and allows us to select the most discriminative non-motor variables to create potential tools for PD screening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative condition after Alzheimer’s disease and affects up to 1% of the population above 60 years [1]. Cardinal motor symptoms such as bradykinesia, rigidity, and resting tremor are essential for PD diagnosis. These motor features emerge when approximately 50% of dopaminergic cells in the substantia nigra have degenerated [2, 3] and 70% of striatum dopaminergic synapses are lost [4]. Therefore, the clinical onset of PD is insidious, and by the time of diagnosis the development of brain pathology is in advanced stages.
Early and accurate detection of PD is crucial for successful outcomes of disease-modifying therapies to slow-down—or even halt—disease progression. Towards this end, clinical features predating motor symptoms might be useful. It is increasingly recognized that non-motor manifestations, including olfactory dysfunction, autonomic symptoms, sleep disorders, visual impairment, cognitive decline or depressive symptoms, not only accompany but usually precede the onset of motor features [5, 6]. This premotor or prodromal phase in PD lasts between 5 to 20 years, and there is an increasing interest in using this array of premotor manifestations to identify PD patients at very early stages.
In the last few decades, machine learning techniques are being increasingly applied for the early diagnosis of PD. This has led to a substantial improvement in prediction accuracy of PD by means of multiple data modalities, including handwritten patterns [7, 8], voice and speech signals [9, 10], different neuroimaging techniques [11,12,13,14,15,16,17,18,19] or biofluids [20, 21]. However, there is a scarcity of studies using machine learning approaches for PD diagnosis based on a constellation of non-motor symptoms.
According to the recent review of works applying machine learning for diagnosis of PD [22], out of 209 publications, 168 focused in discriminating PD patients from controls, but only two reported results obtained using just non-motor symptoms. Prashanth et al. [23] used Support Vector Machine (SVM) algorithm with different kernels. Then, Mabrouk et al. [24] analyzed five machine learning models and compared non-motor features results with results obtained combining DatSCAN single-photon emission computed tomography (SPECT) with motor features.
The rest of the previous works that included non-motor symptoms in their classification algorithms, combined these manifestations with other clinical, imaging and/or biofluid biomarkers [25,26,27,28,29,30]. For example, Prashant et al. [28] obtained high classification accuracy of PD patients and controls with Support Vector Machine (SVM) algorithm using non-motor symptoms, cerebrospinal fluid (CSF) markers and dopamine transporter imaging using SPECT. The combination of non-motor symptoms with other diagnosis modalities might mask the potential classification performance of non-motor symptoms by themselves. A combination of non-motor symptoms with other clinical imaging and/or biofluid biomarkers was also used in some other works pursuing different objectives as PD subtyping [31] or prediction of symptoms of depression [32]. Armañanzas et al. [33] applied classification algorithms exclusively to non-motor symptoms, but their goal was to assess the severity of the disease in PD patients.
Therefore, very few studys have applied classification algorithms exclusively to non-motor symptoms with the goal of discriminating PD patients from controls. Relying on this alternative strategy to detect PD in preclinical stages poses one important advantage over classification algorithms combining multiple data modalities: non-motor symptoms can be used for population PD screening as clinical scales measuring these manifestations are readily available and can be easily implemented as ancillary tests in primary care in contrast to dopaminergic neuroimaging and cerebrospinal fluid measurements, that are usually restricted to specialized hospital care. High diagnostic accuracy of non-motor symptoms for PD diagnosis may encourage the establishment of machine learning algorithms in clinical settings to assist general practitioners in decision making. In addition, building interpretable and/or explainable models will contribute to boost their impact on the healthcare system [34].
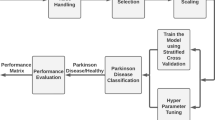
In this work, we selected a combination of clinical scales that measure non-motor features, including depressive symptoms, olfactory function, autonomic symptoms, visuospatial abilities and cognitive outcomes. We evaluated the performance of 9 distinct algorithms for the PD/control (binary) classification using our own cross-sectional database, including 59 PD patients and 37 controls (Biocruces database). Due to the small size of the latter database, we gathered the baseline measurements of Parkinson’s Progression Markers Initiative (PPMI) database and we selected the same or equivalent parameters to those from Biocruces database, which allowed us to additionally include 490 PD patients and 197 controls to our data. Accordingly, we analysed whether the inclusion of these data from a different source could be beneficial to our goals. Two levels of databases were created from each database with increasing granularity: a first level database with total scores of each questionnaire and test (feature per test, FPT) and a second level database that included the individual items of each questionnaire and test (feature per item, FPI). Each level included demographic variables and class (PD or control), as well.
The current work had 2 main objectives. On the one hand, our goal was to assess the ability of well-known machine learning algorithms to distinguish PD patients from control subjects in our database using just demographic data and non-motor manifestations. This goal was complemented with more specific questions: (a) which was the best learning algorithm for this task, (b) which were the most relevant non-motor parameters (features), and (c) could these features produce simple and effective rules? On the other hand, we wanted to evaluate whether data from several clinical studies could be compatible enough so their combination could improve the results achievable separately.
The paper proceeds describing, in Sect. 2, the type of data and tests used in this work, the characteristics of the generated databases, the machine learning methods and the evaluation used for the experiments. In Sect. 3, we explain the results of the performed experiments. In Sect. 4, a comparison with other studies is made and the limitations of the work are explained, while we conclude with some final remarks in Sect. 5.
2 Methodology
2.1 Biocruces database
This database contained information from 59 patients with PD and 37 controls. Participants were recruited and evaluated between the years 2015 and 2018 through the Department of Neurology at Cruces University Hospital and the Biscay PD Association (ASPARBI). All patients fulfilled Parkinson’s UK Brain Bank criteria for the diagnosis of PD. All patients were studied in an on-medication condition to complete all study assessments. The study protocol was approved by the regional Basque Clinical Research Ethics Committee. All participants gave written informed consent prior to their participation in the study, in accordance with the tenets of the Declaration of Helsinki.
The Biocruces database consisted of demographic data and a collection of non-motor clinical outcomes from several questionnaires and psychophysical tests. Assessments were completed on a single occasion (cross-sectional data) by the two neurologists and the two neuropsychologists of the group who were experts in PD. Demographic and general clinical attributes included: gender (GENDER), years of education (EDUCYRS), dominant hand (HANDED), age (AGE) and the class itself (PD or control). The demographic and clinical characteristics of the healthy controls and PD patients included in the Biocruces database are shown in Table 1.
On the other hand, non-motor data included information from neuropsychological tests (Symbol Digit Modalities Test, Benton Judgment of Line Orientation Test, Montreal Cognitive Assessment and Hopkins Verbal Learning Test), from an olfaction test (Brief Smell Identification Test), from a questionnaire on neuropsychiatric symptoms (Geriatric Depression Scale) and autonomic manifestations (SCOPA-AUT). Below we will explain in more detail the content of the aforementioned tests and questionnaires:
-
Geriatric Depression Scale (GDS) [35] is a self-reported 15-item questionnaire for assessing the degree of depression in older adults. Scores of 0-4 are considered normal, depending on age, education, and complaints; 5-8 indicate mild depression; 9-11 indicate moderate depression; and 12-15 indicate severe depression.
-
SCOPA-AUT [36] is a questionnaire that consists of 25 items for assessing the autonomic nervous system. The dysfunction of the following components of the autonomic system are concretely evaluated: gastrointestinal (7 items), urinary (6 items), cardiovascular (3 items), thermoregulatory (4 items), pupillomotor (1 item), and sexual (2 items for men and 2 items for women).
-
Brief Smell Identification Test (BSIT) [37] is used to clinically quantify olfactory deficits, and it includes 12-items. Each item is a single odor that individuals need to identify among 4 options that are provided to test their olfactory function. BSIT is an abbreviated version of University of Pennsylvania Smell Identification Test (UPSIT) [38] test, a comprehensive 40-item test that is divided in 4 booklets of 10-items.
-
Symbol Digit Modalities Test (SDMT) [39] is a simple, fast, and economic test to detect cognitive impairment. It is a paper-pencil measure and consists of substituting digits for abstract symbols using a reference key. The completion of this test requires attention, perceptual speed, motor speed, and visual scanning.
-
Benton Judgment of Line Orientation Test (BJLOT) [40] is a standardized measure of visuospatial abilities. The test measures a person’s ability to match the angle and orientation of lines in space. The complete test has 30 items, but short forms have also been created. Biocruces database includes data from the complete version.
-
Montreal Cognitive Assessment (MoCA) [41] is a neuropsychological test administered by professionals to test the general cognitive abilities of the subject. The score ranges from 0 to 30 points, and scores below 26 are compatible with mild cognitive impairment. It assesses several cognitive domains: short-term memory, spatio temporal reasoning skills, executive functions, attention, concentration and working memory, language, abstraction, reasoning and orientation to time and place.
-
Hopkins Verbal Learning Test (HVLT) [42] is a test of verbal learning and memory. The test consists of three trials of free-recall of a semantically categorized 12-item list, followed by yes/no recognition. Approximately 20–25 min later, a delayed recall trial and a recognition trial are completed. The delayed recall requires free recall of any words remembered. The recognition trial is composed of 24 words, including the 12 target words and 12 false-positives, 6 semantically related, and 6 semantically unrelated words. The revised version (HVLT-R) is more recent and offers six alternate forms. The latter was used in Biocruces database.
We generated 2 versions of the Biocruces database: a first one where each feature represented a single question or test item (feature per item, FPI) and a second one where a single feature was created for each complete questionnaire or test by summing up the answers of its items (feature per test, FPT). The FPI database consisted of 120 variables while the FPT had 14 variables. The first and third columns in Table 2 show the variables in each database and the corresponding equivalencies. Demographic attributes are equal in both databases.
2.2 PPMI database and the combination of Biocruces and PPMI databases
Due to the small sample size of the Biocruces database, we explored the potential benefit of adding data from other studies. For this purpose, we used data from the Parkinson’s Progression Markers Initiative (PPMI),Footnote 1 getting what we called the Bio+PPMI database.
PPMI [43] is a landmark, multicenter, longitudinal study that aims to identify biomarkers for the progression of PD to improve therapeutic and etiological research. The study is a public-private partnership funded by The Michael J. Fox Foundation for Parkinson’s Research (MJFF).Footnote 2 The recruited PD patients form PPMI had a disease duration of 3 years or less and were drug-naïve (early PD patients) at study inclusion.
PPMI has collected longitudinal data from more than 1400 individuals at 33 clinical sites in 11 countries. It began in 2010 but is now recruiting a larger and more diverse group of individuals, including de novo PD, control volunteers and at-risk populations.
The PPMI database has many more parameters than those mentioned above for Biocruces database, thus for the present work we exclusively selected those variables from PPMI database that were also present in Biocruces database. The exported subject data from PPMI database included two cohorts: 490 idiopathic PD patients and 197 controls.
In order to conduct the integration of the Biocruces and PPMI databases, we carried out a unification process transforming several features. First, regarding demographic data, in the PPMI database GENDER takes 3 values (women without reproductive capacity, women with reproductive capacity and men) and just two in the Biocruces database (women and men). Only two possible values were used, merging both women categories from the PPMI database.
In addition, the PPMI study uses the UPSIT instead of BSIT. In order to make the transformation from the UPSIT to the BSIT, we took into account the work of Lawton et al. [44]. This transformation could only be performed for the FPT variables, preventing the compatibility of both databases in the FPI version.
Finally, the version of the BJLOT test was also different in the two databases. PPMI had the abbreviated version of 15 items and the Biocruces database had the complete set of 30 items. This is why the FPT variable of the BJLOT test (BJLOT_total) was divided by two in the Biocruces database.
The demographic and clinical characteristics of the healthy controls and PD patients from the PPMI study are shown in Table 1 together with those from the Biocruces database. When comparing the characteristics of the subjects from both databases, the mean age of the controls in the PPMI database was higher and the proportion of women in PPMI was lower. Differences in age and sex between PD patients from Biocruces and PPMI were more discrete. PD patients from the Biocruces database had slightly worse overall cognitive performance than those from PPMI and more depressive symptoms (GDS), which might be explained by differences in disease duration. Similarly, the motor disability in PD patients from Biocruces was larger.
2.3 Pre-processing of Biocruces and PPMI databases
After unifying both databases, data was pre-processed by removing missing values, outliers and applying rescaling techniques when necessary.
Regarding the SCOPA-AUT test, only questions 1 to 21 were used, due to the large number of missing values in items 22 to 26 in Biocruces database. The missing data from these questions were imputed using the feature mean of each class. Following the pre-processing of the Biocruces database, 5 subjects were removed from the database due to missing data. After such corrections, Biocruces database consisted of 96 individuals, of which 37 were control subjects and 59 PD patients.
Regarding the PPMI database, samples with more than 20% missing values were eliminated, removing a total of 123 subjects. The rest of the missing values were imputed with the feature mean. A total of 687 samples were used, 197 being control subjects and 490 PD patients.
In both Biocruces and PPMI databases, for the questions related to urinary problems in the SCOPA-AUT questionnaire, there is an alternative answer to specify that the subject uses a catheter and, therefore, those questions did not apply. The remaining answers are scaled from 0 (“never”) to 3 (“often”), so we labeled the alternative answer as 4, since it implies urinary problems.
Finally, we observed that the possible range of values varied considerably among different variables. Therefore, we applied two normalisation techniques when it was required by the learning algorithm: one-hot encoding was applied to categorical features, while quantitative ones were normalised by min-max normalisation. This way, all variables took a value in the [0,1] range.
2.4 Feature selection
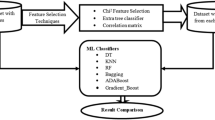
After pre-processing both databases, we applied a feature selection process to identify the most relevant features. This technique was applied to the FPI and FPT versions of both Biocruces and PPMI databases, and also to the database obtained after merging both databases (i.e., Bio+PPMI). A feature selection method was used for two main reasons. On the one hand, one of the aims of this work was to identify the most significant variables or biomarkers to differentiate PD patients from controls. On the other hand, feature selections would help simplifying models, making the information easier to acquire and interpret, and also reducing training times and overfitting probabilities.
Among the feature selection strategies, we decided to use filter methods because they have the lowest computational cost and they are not tuned to a specific type of prediction model [45] and, consequently, can be combined with different classifiers.
We used the multivariate method called Correlated Feature Selection (CFS) [46] because it is able to deal with redundant, duplicated and correlated features. The CFS method was developed by Hall and Smith and searches for subsets that are correlated with the class but independent of each other. The algorithm assumes that features that are irrelevant have a low correlation with the class, so they do not have to be included in the subsets. In addition, they examine excessive features, as these are often correlated with one of the other attributes.
In order to evaluate the subset S of k features, the following formula was used:
where \(\overline{r_{cf}}\) is the average correlation value between the class and the features, and \(\overline{r_{ff}}\) is the average correlation value between all pairs of features.
We tried other filter method such as Fast Correlation Based Filter [47], but the latter obtained less stable subsets, as they depended on the search algorithm.
2.5 Classification algorithms
In this work, the results obtained with 9 supervised machine learning algorithms are compared. To perform a fair comparison, we used their default parameters (see MLP, Bagging and AdaBoost below for the exceptions). All algorithms, except DT and RIPPER, were implemented using the sklearn library in Python. The remaining two were implemented using the Weka software. The main parameters used in the algorithms are described below.
-
Adaptive Boosting (AdaBoost) [48]. We combined 100 DT with the CART algorithm. The weight applied to each classifier at each boosting iteration was 1.
-
Bootstrap aggregating (Bagging) [49]. In order to be comparable with RF, 100 DTs with the CART algorithm were constructed in this work.
-
Decision Tree (DT) [50]. In this project, the C4.5 algorithm [51] was used. The split criterion was the information gain, the minimum number of instances per leaf was 2 and the confidence threshold for pruning was set to 0.25.
-
K-Nearest Neighbors (KNN) [52]. The number of neighbors, K, was set to 5 and the Euclidean distance was used.
-
Multi-Layer Perceptron (MLP) [53]. The architecture had 100 neurons in the hidden layer and, since the system was issuing underfitting warnings with the default value of 200 epochs, we trained it through 500 and 900 epochs in the FPI and FPT databases, respectively. The nonlinear activation function was ReLU and the network was trained using the backpropagation technique [54]. The solver for weight optimization was Adam, with a learning rate of 0.001.
-
Naive Bayes (NB) [55]. The Gaussian Naive Bayes algorithm was used [56], which assumes that the probability of the features is Gaussian.
-
Random Forest (RF) [57]. A set of 100 binary decision trees was used, which were an optimized version of the CART algorithm [58]. The function to measure the quality of a split was Gini impurity, the minimum number of samples required to split an internal node was 2 and the minimum number of samples required to be at a leaf node was 1.
-
Repeated Incremental Pruning to Produce Error Reduction (RIPPER) [59]. The minimal instance weight within a split was 2.
-
Support Vector Machine (SVM) [60]. Radial Basis Function (RBF) kernel with the regularization parameter set to 1 was used.
2.6 Classification performance evaluation
To make a robust estimation, the models were validated using 10 runs of a 10-fold cross-validation [61]. The same seed was used in all classifiers, i.e., all classifiers used the same 10 sub-samples in each run. Performance was mainly evaluated based on accuracy, although F-measure, Precision and Recall were also analyzed.
Statistical significance of the differences in performance was also assessed. First, Fiedman’s Aligned Ranks test [62] was used to test the median equivalence hypothesis. Subsequently, after rejecting the null hypothesis, statistical significances were assessed based on Bayesian statistical tests. The Bayesian approach is based on the subjective interpretation of probability, which considers probability as a degree of belief with respect to uncertainty. We performed the correlated Bayesian test proposed by Corani and Benavoli [63].
2.6.1 Bayesian correlated t-test
The test takes into account that cross-validation on a single database has correlations between training sets, based on the following generative model of the data:
where \(x_{nx1}\) is the vector of accuracy differences, \(1_{nx1}\) is a vector of ones, \(\mu\) is the parameter of interest and \(v \sim MVN(0,\sum _{nxn})\) is a multivariate normal noise with zero mean and covariance matrix \(\sum _{nxn}\) [64].
The posterior distribution can be used to evaluate the probability of one of the algorithms being better than the other or the two algorithms being “practically equivalent”. To do this, we defined that two classifiers were practically equivalent if their mean difference of selected metric was less than a certain value (1% in our case), creating a region of practical equivalence (rope) [65] with the interval [\(-0.01\), 0.01]. Once the rope was defined, the probabilities could be calculated from the posterior:
-
P(left): the integral of the posterior in the interval (\(-\infty\), \(-0.01\)), namely the posterior probability that the mean difference in accuracies is practically negative.
-
P(rope): the integral of the posterior in the interval [\(-0.01\), 0.01], namely the posterior probability that the two classifiers are practically equivalent.
-
P(right): the integral of the posterior in the interval (0.01, \(\infty\)), namely the posterior probability that the mean difference of the accuracies is practically positive.
3 Results
Since one of our goals was to assess the benefits of including additional data to Biocruces database, we designed the experimental work in a progressive way. Firstly, we trained the classification algorithms using exclusively the Biocruces data. Then, as a sanity check of the PPMI database, we replicated the experiment using just the PPMI data. And, finally, we tested whether the combination of Biocruces and PPMI data could be used to improve the results obtained with just the Biocruces database. The following 3 sections describe each of these steps, while Sect. 3.4 is devoted to further analyse comprehensible models aiming to find a set of simple rules that effectively distinguish PD patients from control subjects.
3.1 PD classification in Biocruces database
The first step in the experiment was to apply the CFS feature selection method to the data, resulting in a selection of just 19 features in the FPI version (80% reduction) and 6 in the FPT version (54% reduction), as can be seen in Table 2. The high reduction in the FPI version confirms the expected high correlation among questions in the same questionnaire. The application of the feature selection process duplicated the available database versions, so we applied the 9 supervised machine learning algorithms described in Sect. 2.5 to four versions of the Biocruces database (FPT, FPT+CFS, FPI and FPI+CFS).
Results (accuracy) of supervised machine learning algorithms applied to Biocruces database on correctly discriminating PD patients from healthy controls. Abbreviations: AdaBoost = Adaptive Boosting, Bagging = Bootstrap aggregating, DT = Decision Tree, KNN = K-Nearest Neighbor, MLP = Multi-Layer Perceptron, NB = Naive Bayes, RF = Random Forest, RIPPER = Repeated Incremental Pruning to Produce Error Reduction, SVM = Support Vector Machine
The accuracies of the 9 classification strategies are represented in Fig. 1. The figure shows that most of the algorithms obtained an accuracy greater than 80% to correctly discriminate PD patients from controls, at least for one of the database versions, which proves that many learning strategies achieve quite good detection rates even for small databases. Remarkably, SVM and MLP obtained an accuracy of 86.3% and 84.7%, respectively, although the database version for which the best result was observed did not coincide. A similar trend was detected with the F-Score metric, although the performance of the explanatory models—DT and RIPPER—was a bit lower.
Regarding the database versions, the results didn’t show a clear pattern for all the algorithms. Some algorithms preferred the FPI version while others got better results with the FPT version. However, most of the algorithms showed an improvement in their results when CFS was applied to the data, while just the Bagging algorithm showed a small accuracy and F-score reduction. The performance improvement provided by the feature selection process was especially remarkable for FPI, since 80% of the features from the original database where removed.
3.2 PD classification in PPMI database
As our previous experiment confirmed that good accuracy levels (>80%) could be achieved with features based on non-motor symptoms in a small set of subjects, we proceeded with the replication of the experiment with the equivalent PPMI database. It is worth mentioning that, although the actual goal was to assess the benefit of enriching our data by including samples from other research works, we first replicated the experiment using just the PPMI data to ensure that this new source of information was also well suited for the studied learning algorithms. The obtained results did confirm it.
In fact, many algorithms achieved slightly better accuracy levels compared to the results using Biocruces database. This was an expected finding, as the PPMI database is 7 times larger than the Biocruces one. However, the maximum accuracy level, obtained by the MLP algorithm for the FPI+CFS database version (85.9%), didn’t improve the best value achieved for the Biocruces database (86.3% for SVM with the FPT+CFS database).
The analysis of the differences in performance for the database versions confirmed the conclusions drawn from the Biocruces database. The best database version was algorithm dependent and, for the vast majority, the feature selection process resulted in better accuracy levels. Considering these results and due to the extra benefits a feature selection process provides (e.g., model simplification, speed-up of the training and testing phases), the subsequent analyses were performed using CFS versions of the databases.
3.3 PD classification in combined Biocruces and PPMI database
In this section, we will show the results obtained after enriching our database with the data obtained from the PPMI repository. To that end, we merged both databases, getting the Bio+PPMI database. To get a homogeneous mix, the proportion of subjects from each database was kept constant in the folds of the cross-validation evaluation process.
As explained in Sect. 2, we couldn’t build a FPI version of the merged database due to incompatibilities in data storage format between UPSIT and BSIT in the original databases. This fact, added to our decision of focusing on the feature-reduced versions, let us just the FPT+CFS database version for this experiment. The variables in this database were the following: EDUCYRS, HANDED, AGE, GDS_total, SCAU_total, BJLOT_total, BSIT_total and MoCA_total (see Fig. 2).
Correlated Feature Selection (CFS) of variables from Biocruces database and Bio+PPMI database and PD vs. controls classification performance using 9 supervised machine learning algorithms. * The combination of algorithm and database significantly outperforming other possible combinations (Bayesian tests). Abbreviations: AdaBoost = Adaptive Boosting, Bagging = Bootstrap aggregating, DT = Decision Tree, KNN = K-Nearest Neighbor, MLP = Multi-Layer Perceptron, NB = Naive Bayes, RF = Random Forest, RIPPER = Repeated Incremental Pruning to Produce Error Reduction, SVM = Support Vector Machine
The accuracy and F-score levels of the 9 algorithms are shown in Table 3. The most remarkable finding is that SVM improved the previous results, obtaining 87.5% accuracy. Similarly, MLP improved its accuracy up to 86.9%. Both of them also improved their F-score. In addition, we measured the stability of these estimations and small standard deviation values between 0.4% and 0.7% were computed, proving the reliability of the estimations.
In order to further confirm the validity of the results, we performed a statistical test using the Bayesian tests described in Sect. 2.6.1, after having rejected the equivalence of the medians with the Friedman test (p-value=\(2 \times 10^{-43}\)). For the comparison, 18 cases were defined combining the 9 algorithms with 2 database versions: the FPT+CFS versions of the Biocurces database and the merged database (Bio+PPMI).
The results of the test are summarized in Fig. 3. The green cells show the probability that the algorithm in its row outperforms—by at least 1% of accuracy—the algorithm in its column. The red cells show the opposite. None of the comparisons showed that the most probable result was a tie (i.e., less than 1% difference).
Bayes correlated t-test results between the supervised machine learning algorithms for the two databases (Biocruces and Bio+PPMI). The green colour indicates that the models in the row are statistically better than those in the columns, while the red colour indicates the inverse situation. Abbreviations: Ada. = Adaptive Boosting (AdaBoost), Bag. = Bootstrap aggregating (Bagging), DT = Decision Tree, KNN = K-Nearest Neighbor, MLP = Multi-Layer Perceptron, NB = Naive Bayes, RF = Random Forest, RIP. = Repeated Incremental Pruning to Produce Error Reduction (RIPPER), SVM = Support Vector Machine
The statistical tests mostly confirmed the results given by the mean accuracy values shown in the previous figures and tables. The 3 pairs made up by SVM/Bio+PPMI, MLP/Bio+PPMI and SVM/Biocruces outperformed any other combination with high probability. In particular, SVM/Bio+PPMI did it with 80% or higher probability. It is also remarkable the performance of the SVM algorithm when trained with the small database of Biocruces database.
Considering that these 3 options (SVM/Bio+PPMI, MLP/Bio+PPMI and SVM/Biocruces) are the best ones regarding the accuracy, we analyzed their behavior in more specific situations. On the one hand, we estimated the precision and recall metrics, since a high recall can be preferable in widely applicable screening studies. On the other hand, we estimated the accuracy of the algorithms splitting the test data by the gender variable, since it is well-known that PD’s characteristics often depend on gender and consequently, medical applications designed without taking this aspect into account might generate sub-optimal results as well as discriminatory outcomes [66].
The results, shown in Table 4, suggest that the data enrichment particularly improved the recall values. In this sense, the differences between SVM and MLP were negligible whereas SVM trained with the Biocruces database showed higher precision at the expense of recall reduction. Regarding the gender-specific accuracy, we found significant differences in some of the databases. SVM trained with the Biocruces database was biased towards men subjects while SVM and MLP trained with the merged database showed the opposite bias. These findings suggest that future studies will benefit from generating gender-specific models.
3.4 Comprehensible models for PD detection
In the biomedical context, the interpretability and applicability of classification models in the clinical practice is of utmost importance [34]. In our work, we observed that the rule-based RIPPER algorithm obtained competitive results, since its accuracy was 84.4%, and this result encouraged us to further analyze the rules proposed by the algorithm.
First of all, accepting the high explaining capacity of simple rules, we studied how RIPPER’s accuracy evolved while we forced it to produce simpler rules. To that end, we adjusted the parameter, named N, that controls the minimum number of instances covered by each rule. The default value of N = 2 was replaced by 10, 30 and 50.
The experiments showed that the accuracy was not reduced while the N parameter value was increased. In fact, when just the Biocruces database was used, the accuracy even increased from 81 to 83% when N was changed from 2 to 10. This could be expected since simpler models usually perform better in small databases. However, high N values produced trivial rules due to the small size of the Biocruces database.
The rules generated by RIPPER for the Biocruces database (N=10) and the Bio+PPMI database (N=50) were very similar; they just differed in the threshold proposed for the BSIT test. The first two rows in Table 5 show these rules and their performance metrics. It is remarkable how few simple rules could provide such a good classification performance.
Encouraged by these results, we also applied the RIPPER algorithm with N=10 for the Biocruces FPI+CFS database version (remember that no merged database exists for the FPI version). The accuracy wasn’t reduced compared to the N=2 rules, although it was slightly lower than the results for FPT (80.9%). Nevertheless, the resulting rule shown in the last row of Table 5 was comprised of just two items, where SCAU2 corresponds to the question “In the past month, has saliva dribbled out of your mouth?” and BSIT6 to the rose smell identification, suggesting that an extremely simple rule can be enough for a preliminary patient screening.
4 Discussion
Currently, few works exist that leverage the power of machine learning algorithms to detect PD based on non-motor symptoms and they are supported by more complex data such as clinical images and biofluid biomarkers. Other previous studies have used individual non-motor features of PD to classify PD patients with different objectives. For example, it has been observed that hyposmia has a positive predictive value of 40% in non-PD individuals [67,68,69,70] and predicts the early conversion to PD of patients in prodromal phases [68, 71]. A prospective study also observed 10% risk of conversion of disease-free PD-relatives with hyposmia to symptomatic PD at 2 years [72]. Gastrointestinal disturbances are the most frequent autonomic manifestations of PD patients. Particularly constipation is the most common gastrointestinal symptoms in prodromal PD [73]. However, none of these symptoms present sufficient sensitivity by itself that can be used for screening. As far as we know, only two previous publications have used exclusively non-motor clinical scales and questionnaires to classify PD patients based on machine learning approaches. On one hand, Mabrouk et al. [24] obtained the best accuracy of 82.2% with KNN algorithm. On the other hand, Prashanth et al. [23] got an accuracy of 85.48% using SVM with RBF kernel. Both works included UPSIT, one of the tests coming out to be more discriminant in our work, but combined with different tests. With regard to performance, our best results for PPMI – the database used in both works – is similar, while Biocruces results improved them. The other work we identified using only non-motor features, Armañanzas et al. [33] was aimed to identify non-motor features associated with the severity of motor manifestations, which differs from the objective of our work, in which we seek to identify non-motor characteristics with more accuracy for the early diagnosis of PD versus controls.
In our work, the results obtained with the rule-based RIPPER algorithm showed that even very simple rules based on two single items could achieve good PD detection rates—80.9% in Biocruces database—when trying to discriminate controls from PD. All these results suggest that a hierarchical screening strategy could be designed. This hierarchical screening could begin with just a few questions and tests—such as the identification of a single smell, as suggested by RIPPER—, so it could be universally adopted with little effort. This initial step should filter the subjects less prone to be affected by the PD and redirect the rest to more specific questionnaire and tests; for instance, to the complete SCOPA-AUT and BSIT tests, which formed a rule that correctly classified 84.4% of the subjects in our database. In the final steps, more complex classifiers, such as the SVM model, that achieved an accuracy of 87.5% in our data, could be used.
Hopefully, the steps in the hierarchical screening should begin with simple rules obtaining very high recall values, followed by progressively more complex models with increasingly higher precision. We claim that such a hierarchical approach will drastically improve early detection of PD cases. Obviously, the design of such a strategy requires further research in order to find the best models for each step of the process. To that end, we consider of capital importance to enlarge the available databases, so the learning algorithms can get even more accurate models. Therefore, we welcome all the works aimed at obtaining more data from both, PD patients and control subjects.
One of the main limitations of this study was that it was focused on the classification of controls (that is, without motor disorders suggestive of PD) compared to PD patients with established motor manifestations. Two of the main clinical challenges in PD are the early diagnosis of the disease, even in pre-motor phases, and the differential diagnosis of PD with other conditions that present with tremor, such as neurodegenerative parkinsonism (e.g., multistemic atrophy, progressive supranuclear palsy or Lewy body dementia), vascular parkinsonism, parkinsonism induced by drugs or essential tremor. Although 94% of the patients included in this study were patients with early PD (\(<5\) years duration), their disease was already established clinically, and their differentiation from controls by early biomarkers does not make practical sense. Thus, the results of this work should be considered exclusively as a methodological proposal based on the analysis of non-motor parameters with machine learning to address the mentioned diagnostic challenges in PD. Future studies that analyze non-motor manifestations in a comprehensive way using machine learning to create early diagnostic classification tools should include populations in the pre-motor phase at high risk of developing PD, such as some carriers of genetic mutations or patients with idiopathic REM sleep disorder. Another limitation of the present project is that we have not included some non-motor features relevant to PD, such as the presence of sleep disorders, visual manifestations, neuropsychiatric disorders such as psychotic symptoms or impulse control disorder. Given the potential relevance of these characteristics as early phenomena in PD, future work should also integrate them to make a more comprehensive and exhaustive analysis of the spectrum of non-motor symptoms in PD.
5 Conclusions
It is well established that non-motor manifestations of PD are early and can be quantified daily clinical practice with minimal. Here, we evaluated the performance of 9 distinct machine learning algorithms for discriminating idiopathic PD patients from controls using a wide collection of non-motor clinical PD features. For that, we used information from two databases: Biocruces database (with 59 PD patients and 37 controls) and PPMI database (490 PD and 197 controls). In addition, we evaluated whether the combination of data from such databases could improve the results of machine learning classification compared to those achievable by each database separately.
As main results, we conclude that classifiers induced from non-motor based features are able to discriminate between PD patients and control. We observed that Support Vector Machine (SVM) and Multi-Layer Perceptron (MLP) were the two machine learning algorithms that had the best performance to differentiate PD from controls, with a best accuracy of 87.5% and 86.9%, respectively. Likewise, we observed that the use of correlation feature selection improved the discriminating capacity of the machine learning algorithms.
In order to favor the interpretability and applicability of our tested classification models in the clinical practice, Repeated Incremental Pruning to Produce Error Reduction (RIPPER) machine learning algorithm provided a simple rule based on two non-motor symptoms – SCAU_total (total autonomic manifestations) and BSIT_total (total olfactory dysfunction)–with good accuracy (84.4%), precision (78.1%) and recall (80.8%).
It is important to note that in this kind of studies the PD patients are overrepresented and, hence, the distribution of the training data doesn’t match the test data distribution if applied to a wide segment of the population. As a consequence, specific adaptations will probably be needed in order to properly estimate the model’s performance when applied in such contexts. Moreover, it is important to notice that gender differences exist in non-motor symptoms. Therefore, any clinical implementation effort of classification algorithms for differentiating PD patients from controls, should be predated by gender-specific generation of simple rules.
As a positive final remark, we would like to highlight that the parameters of the learning algorithms weren’t tuned to get the best possible results with our data. This means that there is still room for improvement and that automatic parameter optimization systems could be used to improve the results presented in this article.
References
Lai BC, Tsui JK (2001) Epidemiology of parkinson’s disease. B C Med J 43(3):133–137
Hilker R, Schweitzer K, Coburger S, Ghaemi M, Weisenbach S, Jacobs AH, Rudolf J, Herholz K, Heiss W-D (2005) Nonlinear progression of parkinson disease as determined by serial positron emission tomographic imaging of striatal fluorodopa f 18 activity. Archives Neurol 62(3):378–382
Marek K, Jennings D (2009) Can we image premotor parkinson disease? Neurology 72(7 Supplement 2):21–26
Wüllner U, Pakzaban P, Brownell A-L, Hantraye P, Burns L, Shoup T, Elmaleh D, Petto AJ, Spealman RD, Brownell GL et al (1994) Dopamine terminal loss and onset of motor symptoms in mptp-treated monkeys: a positron emission tomography study with 11c-cft. Exp Neurol 126(2):305–309
Poewe W (2008) Non-motor symptoms in Parkinson’s disease. Eur J Neurol 15:14–20
Murueta-Goyena A, Andikoetxea A, Gómez-Esteban JC, Gabilondo I (2019) Contribution of the gabaergic system to non-motor manifestations in premotor and early stages of Parkinson’s disease. Front Pharmacol 10:1294
Adams WR (2017) High-accuracy detection of early parkinson’s disease using multiple characteristics of finger movement while typing. PloS One 12(11):0188226
Drotár P, Mekyska J, Rektorová I, Masarová L, Smékal Z, Faundez-Zanuy M (2014) Decision support framework for parkinson’s disease based on novel handwriting markers. IEEE Transactions Neural Syst Rehabil Eng 23(3):508–516
Sakar BE, Isenkul ME, Sakar CO, Sertbas A, Gurgen F, Delil S, Apaydin H, Kursun O (2013) Collection and analysis of a Parkinson speech dataset with multiple types of sound recordings. IEEE J Biomed Health Informatics 17(4):828–834
Ma H, Tan T, Zhou H, Gao T (2016) Support vector machine-recursive feature elimination for the diagnosis of parkinson disease based on speech analysis In: 2016 Seventh International Conference on Intelligent Control and Information Processing (ICICIP), pp 34–40 IEEE
Segovia F, Górriz JM, Ramírez J, Martínez-Murcia FJ, Castillo-Barnes D (2019) Assisted diagnosis of parkinsonism based on the striatal morphology. Int J Neural Syst 29(09):1950011
Nunes A, Silva G, Duque C, Januário C, Santana I, Ambrósio AF, Castelo-Branco M, Bernardes R (2019) Retinal texture biomarkers may help to discriminate between Alzheimer’s, Parkinson’s, and healthy controls. PloS One 14(6):0218826
Fernández-Carmona A, Olivencia-Peña L, Yuste-Ossorio M, Peñas-Maldonado L et al (2018) Ineffective cough and mechanical mucociliary clearance techniques. Medicina Intensiva (English Edition) 42(1):50–59
Nuvoli S, Spanu A, Fravolini ML, Bianconi F, Cascianelli S, Madeddu G, Palumbo B (2020) [123i] metaiodobenzylguanidine (mibg) cardiac scintigraphy and automated classification techniques in Parkinsonian disorders. Mol Imag Biol 22(3):703–710
Adeli E, Shi F, An L, Wee C-Y, Wu G, Wang T, Shen D (2016) Joint feature-sample selection and robust diagnosis of parkinson’s disease from mri data. NeuroImage 141:206–219
Amoroso N, La Rocca M, Monaco A, Bellotti R, Tangaro S (2018) Complex networks reveal early mri markers of Parkinson’s disease. Med Image Anal 48:12–24
Ariz M, Abad RC, Castellanos G, Martínez M, Muñoz-Barrutia A, Fernández-Seara MA, Pastor P, Pastor MA, Ortiz-de-Solórzano C (2018) Dynamic atlas-based segmentation and quantification of neuromelanin-rich brainstem structures in Parkinson disease. IEEE Transactions Med Imag 38(3):813–823
Rana B, Juneja A, Saxena M, Gudwani S, Kumaran SS, Agrawal R, Behari M (2015) Regions-of-interest based automated diagnosis of Parkinson’s disease using t1-weighted mri. Expert Syst Appl 42(9):4506–4516
Shinde S, Prasad S, Saboo Y, Kaushick R, Saini J, Pal PK, Ingalhalikar M (2019) Predictive markers for parkinson’s disease using deep neural nets on neuromelanin sensitive mri. NeuroImage Clinic 22:101748
Váradi C, Nehéz K, Hornyák O, Viskolcz B, Bones J (2019) Serum n-glycosylation in parkinson’s disease: a novel approach for potential alterations. Molecules 24(12):2220
Maass F, Michalke B, Willkommen D, Leha A, Schulte C, Tönges L, Mollenhauer B, Trenkwalder C, Rückamp D, Börger M et al (2020) Elemental fingerprint: reassessment of a cerebrospinal fluid biomarker for Parkinson’s disease. Neurobiol Dis 134:104677
Mei J, Desrosiers C, Frasnelli J (2021) Machine learning for the diagnosis of Parkinson’s disease: a review of literature. Front Aging Neurosci 13:184
Prashanth R, Roy SD, Mandal PK, Ghosh S (2014) Parkinson’s disease detection using olfactory loss and rem sleep disorder features In: 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp 5764–5767 IEEE
Mabrouk R, Chikhaoui B, Bentabet L (2018) Machine learning based classification using clinical and datscan spect imaging features: a study on parkinson’s disease and swedd. IEEE Transactions Radiat Plasma Med Sci 3(2):170–177
Challa KNR, Pagolu VS, Panda G, Majhi B (2016) An improved approach for prediction of parkinson’s disease using machine learning techniques In: 2016 International Conference on signal processing, communication, power and embedded system (SCOPES), pp 1446–1451 IEEE
Dhami DS, Soni A, Page D, Natarajan S (2017) Identifying parkinson’s patients: a functional gradient boosting approach In: Conference on Artificial Intelligence in Medicine in Europe, pp. 332–337 Springer
Dinov ID, Heavner B, Tang M, Glusman G, Chard K, Darcy M, Madduri R, Pa J, Spino C, Kesselman C et al (2016) Predictive big data analytics: a study of parkinson’s disease using large, complex, heterogeneous, incongruent, multi-source and incomplete observations. PloS One 11(8):0157077
Prashanth R, Roy SD, Mandal PK, Ghosh S (2016) High-accuracy detection of early parkinson’s disease through multimodal features and machine learning. Int J Med Informatics 90:13–21
Prince J, Andreotti F, De Vos M (2018) Multi-source ensemble learning for the remote prediction of Parkinson’s disease in the presence of source-wise missing data. IEEE Transactions Biomed Eng 66(5):1402–1411
Leger C, Herbert M, DeSouza JF (2020) Non-motor clinical and biomarker predictors enable high cross-validated accuracy detection of early pd but lesser cross-validated accuracy detection of scans without evidence of dopaminergic deficit. Front Neurol 11:364
Zhang X, Chou J, Liang J, Xiao C, Zhao Y, Sarva H, Henchcliffe C, Wang F (2019) Data-driven subtyping of parkinson’s disease using longitudinal clinical records: a cohort study. Scientific Rep 9(1):797. https://doi.org/10.1038/s41598-018-37545-z
Gu S-C, Zhou J, Yuan C-X, Ye Q (2020) Personalized prediction of depression in patients with newly diagnosed Parkinson’s disease: a prospective cohort study. J Affect Disord 268:118–126. https://doi.org/10.1016/j.jad.2020.02.046
Armañanzas R, Bielza C, Chaudhuri KR, Martinez-Martin P, Larrañaga P (2013) Unveiling relevant non-motor Parkinson’s disease severity symptoms using a machine learning approach. Artif Intell Med 58(3):195–202. https://doi.org/10.1016/j.artmed.2013.04.002
Vellido A (2020) The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Comput Appl 32(24):18069–18083. https://doi.org/10.1007/s00521-019-04051-w
Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, Leirer VO (1982) Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res 17(1):37–49
Visser M, Marinus J, Stiggelbout AM, Van Hilten JJ (2004) Assessment of autonomic dysfunction in Parkinson’s disease: the scopa-aut. Mo Disord Off J Mov Disord Soc 19(11):1306–1312
Doty RL, Marcus A, William Lee W (1996) Development of the 12-item cross-cultural smell identification test (cc-sit). Laryngoscope 106(3):353–356
Doty RL, Shaman P, Kimmelman CP, Dann MS (1984) University of pennsylvania smell identification test: a rapid quantitative olfactory function test for the clinic. Laryngoscope 94(2):176–178
Smith A (1968) The symbol digit modalities test: a neutopsychologic test for economic screening of learning and other cerebral disorders Learning Disorders 3, 83-91
Benton AL, Varney NR, Hamsher Kd (1978) Visuospatial judgment:a clinical test. Archives Neurol 35(6):364–367
Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H (2005) The montreal cognitive assessment, moca: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 53(4):695–699
Benedict RH, Schretlen D, Groninger L, Brandt J (1998) Hopkins verbal learning test-revised: normative data and analysis of inter-form and test-retest reliability. Clin Neuropsychologist 12(1):43–55
Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S et al (2011) The Parkinson progression marker initiative (ppmi). Progress Neurobiol 95(4):629–635
Lawton M, Hu MT, Baig F, Ruffmann C, Barron E, Swallow DM, Malek N, Grosset KA, Bajaj N, Barker RA et al (2016) Equating scores of the university of pennsylvania smell identification test and sniffin’sticks test in patients with parkinson’s disease. Parkinsonism Relat Disord 33:96–101
Zhang Y, Li S, Wang T, Zhang Z (2013) Divergence-based feature selection for separate classes. Neurocomputing 101:32–42
Hall MA (1999) Correlation-based feature selection for machine learning. Department of Computer Science, Waikato University, PhD Thesis, New Zealand
Yu L, Liu H (2003) Feature selection for high-dimensional data: a fast correlation-based filter solution In: Proceedings of the 20th International Conference on Machine Learning (ICML-03), pp 856–863
Schapire RE (2013) Explaining adaboost. In: Schölkopf B, Luo Z, Vovk V (eds) Empirical Inference: Festschrift in Honor of Vladimir N. Vapnik. Springer, Berlin, Heidelberg, pp 37–52. https://doi.org/10.1007/978-3-642-41136-6_5
Breiman L (1996) Bagging predictors. Mach Learn 24(2):123–140
Xu M, Watanachaturaporn P, Varshney PK, Arora MK (2005) Decision tree regression for soft classification of remote sensing data. Remote Sens Environ 97(3):322–336
Ruggieri S (2002) Efficient c4. 5 [classification algorithm] IEEE transactions on knowledge and data engineering 14(2), 438–444
Altman NS (1992) An introduction to kernel and nearest-neighbor nonparametric regression. Am Statistician 46(3):175–185
Gardner MW, Dorling S (1998) Artificial neural networks (the multilayer perceptron)-a review of applications in the atmospheric sciences. Atmospheric Environ 32(14–15):2627–2636
Leung H, Haykin S (1991) The complex backpropagation algorithm. IEEE Transactions Signal Process 39(9):2101–2104
Zhang H (2004) The optimality of naive bayes. AA 1(2):3
Jahromi AH, Taheri M (2017) A non-parametric mixture of gaussian naive bayes classifiers based on local independent features In: 2017 Artificial Intelligence and Signal Processing Conference (AISP), pp 209–212 IEEE
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Rutkowski L, Jaworski M, Pietruczuk L, Duda P (2014) The cart decision tree for mining data streams. Information Sci 266:1–15
Cohen WW (1995) Fast effective rule induction In: Machine Learning Proceedings 1995, pp 115–123 Elsevier, Amsterdam, The Netherlands
Pisner DA, Schnyer DM (2020) Support vector machine In: Machine Learning, pp 101–121 Elsevier, Amsterdam, The Netherlands
Purushotham S, Tripathy B (2011) Evaluation of classifier models using stratified tenfold cross validation techniques In: International Conference on Computing and Communication Systems, pp 680–690 Springer
García S, Fernández A, Luengo J, Herrera F (2010) Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: experimental analysis of power. Information Sci 180(10):2044–2064
Corani G, Benavoli A (2015) A bayesian approach for comparing cross-validated algorithms on multiple data sets. Mach Learn 100(2):285–304
Benavoli A, Corani G, Demšar J, Zaffalon M (2017) Time for a change: a tutorial for comparing multiple classifiers through bayesian analysis. J Mach Learn Res 18(1):2653–2688
Kruschke J, Liddell T (2015) The bayesian new statistics: two historical trends converge. SSRN Electron J. https://doi.org/10.2139/ssrn.2606016
Cirillo D, Catuara-Solarz S, Morey C, Guney E, Subirats L, Mellino S, Gigante A, Valencia A, Rementeria MJ, Chadha AS, Mavridis N (2020) Sex and gender differences and biases in artificial intelligence for biomedicine and healthcare. NPJ Digital Med 3(1):81. https://doi.org/10.1038/s41746-020-0288-5
Berg D, Postuma RB, Adler CH, Bloem BR, Chan P, Dubois B, Gasser T, Goetz CG, Halliday G, Joseph L et al (2015) Mds research criteria for prodromal Parkinson’s disease. Mov Disord 30(12):1600–1611
Mahlknecht P, Iranzo A, Högl B, Frauscher B, Müller C, Santamaría J, Tolosa E, Serradell M, Mitterling T, Gschliesser V et al (2015) Olfactory dysfunction predicts early transition to a lewy body disease in idiopathic rbd. Neurology 84(7):654–658
Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, Litvan I, Lang AE et al (2015) Mds clinical diagnostic criteria for Parkinson’s disease. Mov Disord 30(12):1591–1601
Boeve BF (2013) Idiopathic rem sleep behaviour disorder in the development of Parkinson’s disease. Lancet Neurol 12(5):469–482
Fereshtehnejad S-M, Montplaisir JY, Pelletier A, Gagnon J-F, Berg D, Postuma RB (2017) Validation of the mds research criteria for prodromal Parkinson’s disease: longitudinal assessment in a rem sleep behavior disorder (rbd) cohort. Mov Disord 32(6):865–873
Ponsen MM, Stoffers D, Booij J, van Eck-Smit BL, Wolters EC, Berendse HW (2004) Idiopathic hyposmia as a preclinical sign of Parkinson’s disease. Annal Neurol Off J Am Neurol Assoc Child Neurol Soc 56(2):173–181
Stirpe P, Hoffman M, Badiali D, Colosimo C (2016) Constipation: an emerging risk factor for Parkinson’s disease? Eur J Neurol 23(11):1606–1613
Acknowledgements
We thank all the participants involved in the study.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was partially funded by the Department of Education, Universities and Research of the Basque Government (ADIAN, IT-980-16); by the Spanish Ministry of Science, Innovation and Universities - National Research Agency and the European Regional Development Fund - ERDF (PhysComp, TIN2017-85409-P), and from the State Research Agency (AEI, Spain) under grant agreement No RED2018-102312-T (IA-Biomed); by Michael J. Fox Foundation [RRIA 2014 (Rapid Response Innovation Awards) Program (Grant ID: 10189)]; by the Instituto de Salud Carlos III through the project “PI14/00679” and “PI16/00005”, the Juan Rodes grant “JR15/00008” (IG) (Co-funded by European Regional Development Fund/European Social Fund - “Investing in your future”); and by the Department of Health of the Basque Government through the projects “2016111009” and “2019111100”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
The study protocol was approved by the regional Basque Clinical Research Ethics Committee.
Consent to participate
All participants gave written informed consent prior to their participation in the study, in accordance with the tenets of the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Martinez-Eguiluz, M., Arbelaitz, O., Gurrutxaga, I. et al. Diagnostic classification of Parkinson’s disease based on non-motor manifestations and machine learning strategies. Neural Comput & Applic 35, 5603–5617 (2023). https://doi.org/10.1007/s00521-022-07256-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07256-8