Abstract
The recognition of stomatological disorders and the classification of dental caries are important areas of biomedicine that can hugely benefit from machine learning tools for the construction of relevant mathematical models. This paper explores the possibility of using reflectivity data to distinguish between healthy tissues and caries by deep learning and multilayer convolutional neural networks. The experimental data set includes more than 700 observations recorded in the stomatology laboratory. For rigor, the results obtained from the deep learning systems are compared with those evaluated for selected sets of features estimated for each observation and classified by a decision tree, support vector machine (SVM), k-nearest neighbor, Bayesian methods, and two-layer neural networks. The classification accuracy obtained for the deep learning systems was 98.1% and 94.4% for data in the signal and spectral domains, respectively, in comparison with an accuracy of 97.2% and 87.2% evaluated by the SVM method. The proposed method conclusively demonstrates how the artificial intelligence and deep learning methodology can contribute to improved diagnosis of dental problem in stomatology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Deep learning methods [37] are transforming the way we approach many engineering and biomedical areas both for image and signal processing [2, 25], recognition of multi-dimensional signal components, and their classification [34]. In particular, artificial intelligence and convolutional neural networks offer important perspectives for the analysis of medical data [8, 39, 41].
When it comes to stomatology, machine learning methods [11] are used for tasks including automatic spatial detection of teeth positioning [3, 5, 9] during oral operations and analysis of periapical radiographs. Specific studies analyze the contrast between sound and lesion areas on occlusal surfaces for different wavelengths of the near-infrared light [23]. Early success has demonstrated that selected computational methods also allow improvements over the standard visual diagnostics. The associated applications include the detection of the dental plaque causing many oral diseases [6, 22] and the analysis of dental caries that are characterized by demineralization of hydroxyapatite crystals and destruction of collagen matter in dental tissues [20].
The need for the early detection of this pathological situation motivates the study of different methods for the examination of tissues, including spectroscopy, radiography, and conductance measurement. Raman spectroscopy [12], different mathematical methods, and neural network models are extensively studied in this connection for the analysis of recorded signals and images as well. The classification of data segments of biomedical signals may be performed by classical clustering methods [4, 32, 33] or by deep learning algorithms [7, 24, 36]. Standard clustering and classification methods, including decision trees (DT), the k-nearest neighbors method (kNM), support vector machine (SVM), Bayesian methods, and two-layer neural network (NN) algorithms, have been successfully used in this area [14, 19, 31] to detect different kinds of disorders.
Deep learning methods based upon multilayer neural networks are often trained over an extensive number of signals. However, the further problems that need to be solved prior to their efficient use include the selection of data sets such that the individual classes have similar sizes [40]. Problems of class imbalance and associated classification issues [21] are often reduced by data augmentation methods to enlarge the extent of the training data sets. This approach was tested also in stomatology [17, 18] for analysis of dental tissue changes and their classification by convolutional neural networks.
The present paper is devoted to the classification of reflectivity data recorded for healthy dental tissues and caries, as outlined in Fig. 1. This approach is often applied as a tool for surface defect analysis [42] and the detection of dental disorders [3, 4]. The goal of the present study is to compare deep learning and standard classification methods in this area, so as to establish the extent to which artificial intelligence methods can be reliably used in early diagnosis of dental disorders. This promises to pave the path for the improvement in treatment outcomes and a reduction in the need for invasive procedures.
2 Methods
2.1 Data acquisition
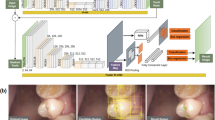
Measurement of the interaction of the light in biological tissues is one type of optical spectroscopy that can be used for the noninvasive or minimally invasive evaluation of tissue pathology [1, 16]. The fundamental blocks of such systems are presented in Fig. 1a. In principle, it is an optical spectroscopy technique whereby a broadband white light is used to illuminate a tissue through an optical fiber probe. The reflected light is captured by the same or separate probe after being subjected to absorption and scattering in the selected tissue area. A multispectral detection system then records the reflection for different wavelengths during each experiment.
Data classification including: a, b the training and testing sets of reflectivity curves of healthy tissues (class A) and tissues with caries (class B), c classification systems using an artificial neural network with five layers based upon deep learning using specific target values for each pattern vector, and d the validation process providing probabilities of estimated classes
Spectroscopy systems in stomatology use a commonly accepted scoring system [27] that is based on the International Caries Detection and Assessment System (ICDAS). The experiments presented here included 741 measurements of healthy and unhealthy tissues, reduced by 6.9% unreliable data to 690 records. This process eliminated data whose difference from the mean curve for each class exceeded the selected threshold. The scoring system used by an experienced stomatologist then associated each measurement \(j=1,2,\ldots ,Q\) out of \(Q=690\) with either class A (422 healthy tissues) or class B (268 caries), with specific probability values. All measurements were randomly divided into two parts that included:
-
1.
The training set (Fig. 2a) of 90% (621) of the observations belonging to classes A (380) and B (241), respectively, for constructing the classification model,
-
2.
The testing set (Fig. 2b) of 10% (69) of observations belonging to classes A (42) and B (27), respectively, for testing the classification model.
The selection of the training and testing sets was performed randomly, to compare the classification accuracy for different groups of observations.
Figure 2a presents the training sets of observations belonging to classes A and B, together with their mean values, which show typical reflectivity plots for healthy tissues and those with dental caries. The shape of these curves is in agreement with other studies [38], pointing out that spectral components less than 570 nm (the blue-green area) are marginal for healthy tissues only.
For each experiment of a specific class, the reflectivity was evaluated and the associated features were estimated. The whole set of experiments was then analyzed by statistical tools and selected classification methods.
2.2 Feature extraction and classification
The data processing presented in Fig. 1b included the analysis of the individual records of reflectivity, evaluation of their mean values, and specification of typical reflectivity curves for each class, as presented in Fig. 2a. Data resampling was then employed, so as to have values on the regular grid of the independent variable. The standard deviation of the differences between each measurement and these typical curves was then used as a measure to extract records affected by gross measurement errors.
The pattern matrix used for the following classification was next defined for the training set, with 90% of the measured values randomly chosen, and the following construction of the pattern matrix columns standing for separate observations and pattern vectors specified by:
-
All values of each reflectivity curve inside the selected wavelength range \(\langle 450, 1550 \rangle \) nm with the sampling period of 1 nm (used for the following deep learning process),
-
Selected features associated with each reflectivity curve estimated both in the time and frequency domains (for the following standard classification).
Target values associated with each column of the pattern matrix were specified by an experienced stomatologist in both cases.
Reflectivity spectrum polynomial approximation (RSA) presenting: a, d a selected reflectivity spectrum (RS) record and its approximation by a polynomial of degree 5, b, e the set of approximate values, and c, f differences between reflectivity values and their approximation for class A and B respectively
The polynomial approximation of each reflectivity curve \(\{s_j(n)\}_{n=0}^{N-1}\) of the selected degree was then applied to each experiment \(j=1,2,\ldots ,Q\) with results presented in Fig. 3a, d for selected sets of each class. Figure 3b, e present these approximations \(\{f_j(n)\}_{n=0}^{N-1}\) for all measurements with differences \(\{d_j(n)\}_{n=0}^{N-1}\) between reflectivity values and their approximations presented in Fig. 3c, f.
The features used for standard classification methods included:
-
1.
The wavelength for the maximal value of reflectivity,
-
2.
The standard deviation of differences between reflectivity values and their polynomial approximations of a selected degree \(M=5\).
Spectrograms of each difference signal \(\{d_j(n)\}_{n=0}^{N-1}\) for both classes were then used to evaluate the relative mean power in frequency ranges \(r_1=\langle f_{c1}, f_{c2} \rangle \) and \(r_2=\langle f_{c3}, f_{c4} \rangle \) that formed spectral domain features for the following classification.
The features of each difference signal \(\{d_j(n)\}_{n=0}^{N-1}\) for \(j=1,2,\ldots ,Q\) were evaluated by the discrete Fourier transform as the relative power p(i, j) estimated both in the frequency range \(r_1\) and \(r_2\) using the following relations:
where \(\Phi _i\) is the set of indices for the range of frequencies \(f_k\! \in r_i \), \(i=1,2\), and \(j=1,2,\ldots ,Q\) for the total number of experiments Q. The pattern matrix \(\mathbf{P}_{2,Q}\) is defined in this way.
Figure 4 presents the distribution of selected couples of features used for the following classification into two classes in the signal and spectral domains. Figure 4a presents the standard deviation of differences between signals and their polynomial approximations versus the wavelength of the maximal value of the reflectivity curve. The distribution of the features evaluated in the spectral domain is presented in Fig. 4b. It presents the relative power of the differences between the signals and their approximations in two selected frequency ranges. The centers of the individual classes and c-multiples of standard deviations for \(c=0.2, 0.5\), and 1 are presented in both cases as well.
The distribution of features for classification into two classes using: a the standard deviation of differences between signals and their polynomial approximations versus the wavelength of the maximal value of the reflectivity curve and b the relative power of differences between signals and their approximations in two selected frequency ranges with centers of individual classes and c-multiples of standard deviations for \(c=0.2, 0.5\) and 1
2.3 Pattern recognition
A supervised learning strategy [10, 28] was used for classification into two classes specified in the target vector \(\mathbf{T}_{S,Q}\) to separate the individual signals into \(S=2\) categories, namely into class A (healthy tissues), and class B (tissues with caries). These classes were defined by an experienced stomatologist for the learning set of signals. To initialize the classification process, the pattern matrix \(\mathbf{P}_{R,Q}\) was constructed with each its column \(j=1,2,\ldots ,Q\) specifying an individual signal in the learning stage.
In the case of the deep learning strategy, no specific features were evaluated and each feature vector associated with signal \(j=1,2,\ldots ,Q\) included all signal values or their transform into the spectral domain. The number R of pattern matrix rows corresponded with the number of signal or spectral values. This process is presented in Fig. 2. The observed signal set was randomly divided into two groups: the first one used for training and the second one for testing, as presented in Fig 2a, b. The classification system presented in Fig. 2c was formed by five layers: the input layer, bidirectional long short-term memory (LSTM) [26] layer, fully connected layer, softmax layer, and the classification layer [13]. The optimization of model parameters was performed by the ADAM method [15].
To compare the results of the deep learning framework with the standard classification methods, two features were evaluated for each signal and the matrix \(\mathbf{P}_{2,Q}\) specified by Eq. (1) was constructed. This kind of information compression enabled both the graphical presentation of feature clusters in the two-dimensional space and the use of standard classification methods. These included the support vector machine, a Bayesian method, the k-nearest neighbor method, and a two-layer neural network. The accuracies and k-fold cross-validation errors of the final models were then used for evaluating the selected classification methods.
The standard classification models used the same set of signals for training of the individual models. The two-layer neural network system was formed by the simplified system presented in Fig. 2c: (1) a fully connected layer of a selected number of elements with the sigmoidal transfer function and (2) the classification layer. The pattern matrix \(\mathbf{P}_{R,Q}\) formed the input for the two-layer neural network to evaluate the values using the following relation:
The network coefficients included the elements of the matrices \(\mathbf{W1}_{S1,R}\), \(\mathbf{W2}_{S,S1}\) and vectors \(\mathbf{b1}_{S1,1}\), \(\mathbf{b2}_{S,1}\). The classification models used the sigmoidal transfer function f1 and the softmax transfer function f2 in the first and the second layer, respectively [35].
The number of output elements corresponded with the number S of classes. The values in each column \(j=1,2,\ldots ,Q\) of the output matrix \(\mathbf{A}_{S,Q}\) defined the predicted probabilities of the belongingness to each class. The row number with the highest probability pointed to the estimated class number for each pattern vector.
During the learning process, the network coefficients were optimized in separate learning epochs and the performance of the classification models was evaluated by the log-loss function, which takes into account the probability that is assigned to the estimation of the target value.
The classification results obtained after the selected number of training epochs were then analyzed by the receiver operating characteristic (ROC) [29, 30]. The learning process evaluated the number of true/false negatives (TN, FN) and true/false positives (TP, FP) both in the negative set (class A: healthy tissue) and positive set (class B: caries). These measures formed the confusion matrix:
The associated performance metrics included:
-
The true rate (A-negative, B-positive)
$$\begin{aligned} \mathrm{TR}(\mathrm{A})= \frac{\mathrm{TN}}{\mathrm{TN}+\mathrm{FP}}, \mathrm{TR}(\mathrm{B})= \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}} \end{aligned}$$(4) -
The predictive value (A-negative, B-positive)
$$\begin{aligned} \mathrm{PV}(\mathrm{A})= \frac{\mathrm{TN}}{\mathrm{TN}+\mathrm{FN}}, \mathrm{PV}(\mathrm{B}) = \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FP}} \end{aligned}$$(5) -
The accuracy value
$$\begin{aligned} \mathrm{ACC}= \frac{\mathrm{TP}+\mathrm{TN}}{\mathrm{TP}+\mathrm{TN}+\mathrm{FP}+\mathrm{FN}} \end{aligned}$$(6)
The k-fold cross-validation method was then employed as a measure of the generalization capabilities of each neural network model.
3 Results
The classification of the reflectivity data of dental tissues into two classes (A: healthy, B: caries) was performed using original data sets acquired in the stomatologic laboratory by the second author. Figure 5 shows the results of the deep learning method, presenting the training accuracy and the loss for different randomly selected initializations of the whole model over ten experiments. The goal of the process was the optimization of the network to maximize the accuracy and to minimize the loss during individual epochs. This optimization was performed by the adaptive moment estimation (ADAM) method [15] in the mathematical and software environment of Matlab2021a.
Results of the deep learning-based classification: training accuracy and loss values for feature matrix constructed by column vectors of a individual signals and b spectral values of their differences related to the polynomial approximation of the fifth degree over 300 training epochs for different selections of training subsets and initial conditions
The optimization process was performed for the deep neural network structure presented in Fig. 2c. The proposed model was constructed as a five-layer neural network for the LSTM layer with 100 units followed by the fully connected layer with two outputs. The initial training rate and gradient threshold were 0.002 and 1, respectively. The optimization process was performed in the proposed incremental mode that allowed modifying the structure and network coefficients during the learning stage. Figure 6 presents the incremental values of the accuracy and the loss in the subsequent learning stages together with their mean values over all values in each stage. This process enabled improving the behavior of the deep neural network used for classification into two classes with the randomly selected training system initializations.
Table 1 presents the accuracy ACC and the loss value LV for ten deep learning experiments and different network initializations presenting the final value (F) after 300 learning epochs and its mean (F10) evaluated from its ten last values. The training was performed for the training set of values presented in Fig. 2a and verification for the different set in Fig. 2b of 10% of experiments randomly chosen from all observations. The mean value of the accuracy and the loss evaluated from the last 10 epochs were 97.96% and 0.06, respectively.
Tables 2 and 3 present the confusion matrix of the classification by the deep learning neural network model for the training and testing sets and the selected experiment. The results based on the signal domain features presented in Table 2 show higher precision both for class A and class B. The sensitivity and specificity are higher for the signal domain pattern matrix and the final accuracy for signal and spectral domain features is 98.1% and 94.4%, respectively.
Classification of healthy tissues and caries for two features evaluated as a, b the wavelength of the maximal reflectivity value and the standard deviation of the differences of reflectivity values and their approximation and c, d the relative power of the difference signal in two frequency bands using a, c the SVM machine and b, d the two-layer neural network
The classification results achieved by the deep neural network were compared with those achieved by the SVM machine, Bayesian method, and the two layer neural network with the sigmoidal and softmax transfer function. Figure 7 presents the classification into two classes (healthy tissues and caries) for two features evaluated as the wavelength of the maximal reflectivity value and the standard deviation of the reflectivity values in the decreasing part of reflectivity using selected classification methods with accuracy (ACC) and cross-validation (CV) errors with class boundaries.
A comparison of the accuracy and cross-validation errors for the classification of stomatological data into two classes by standard methods (support vector, Bayesian, k-nearest neighbor, the two-layer neural network), and the five-layer deep neural network after 300 learning epochs is presented in Table 4. For the given data set, the results of the classifications by the standard classification methods both for the signal and spectral domain features were very close. The classification accuracy for the deep learning systems is 98.1% and 94.4% for data in the signal and spectral domains, respectively, with the loss criterion below 0.01 in both cases. The efficiency of the deep learning is much higher for the spectral domain features with not so well-separated clusters.
4 Conclusion
This paper investigated the effectiveness of selected mathematical methods for the classification of reflectivity data recorded for a total number of 741 dental tissues in the stomatological laboratory. The classification into two categories (healthy tissues and caries) was performed by a deep learning neural network and the ADAM method for model parameter optimization. Its results were compared with classifications by standard methods, which included the support vector machine, a Bayesian method, k-nearest neighbors, and a two-layer neural network. For the deep learning method, no features were evaluated and the complete records formed the input into the classification system. By contrast, in the standard approach to classification based on pattern recognition, feature vectors were estimated from reflectivity data preprocessed by the proposed polynomial approximation.
The results have shown that the deep learning neural network system was able to distinguish caries from healthy tissues without any preprocessing or selection of features, with better accuracy than the standard methods. The classification accuracy was 98.1% and 94.4% for the deep learning system using signal and spectral domain features, respectively. The loss criterion was below 0.01 in both cases.
The deep learning system for signal segment classification has been able to eliminate the process of selecting signal features, which can be sometimes rather complicated and time-consuming. On the other hand, the advantage of the evaluation of features can lie in the deeper understanding of the signal properties both in the time and frequency domain and the possibilities of their visualization. The combination of both approaches to data processing can bring their benefits to obtain a deeper understanding of the physical background of the studied system.
It is expected that further research will be devoted to the deep learning and standard classification systems to benefit from their specific advantages. In the case of deep learning systems, with their complicated structures and with the possibilities of an incremental learning strategy, it will be possible to study the effects of varying the system structure and its coefficients during the learning process. Further studies of the distribution of specific features will enable a deeper data analysis and the use in the clinical environment. In both cases, new machine learning strategies and their implementation for real biomedical data processing and for monitoring physiological functions will be studied as well.
References
Bachmann L, Zezell DM, Ribeiro ADC, Gomes L, Ito AS (2006) Fluorescence spectroscopy of biological tissues—a review. Appl Spectrosc Rev 41:575–590
Cao C, Liu F, Tan H, Song D, Shu W, Li W, Zhou Y, Bo X, Xie Z (2018) Deep learning and its applications in biomedicine. Genomics Proteomics Bioinform 16(1):17–32
Casalegno F, Newton T, Daher R, Abdelaziz M, Lodi-Rizzini A, Schürmann F, Krejci I, Markram H (2019) Caries detection with near-infrared transillumination using deep learning. J Dent Res 98(11):1227–1233
Charvát J, Procházka A, Fričl M, Vyšata O, Himmlová L (2020) Diffuse reflectance spectroscopy in dental caries detection and classification. SIViP 14:1063–1070
Chen H, Zhang K, Lyu P, Li H, Zhang L, Wu J, Lee C (2019) A deep learning approach to automatic teeth detection and numbering based on object detection in dental periapical films. Sci Rep 9:3840:1-3840:11
Chen Y, Stanley K, Att W (2020) Artificial intelligence in dentistry: current applications and future perspectives. Quintessence Gen Dent 51(3):248–257
Fawaz H, Forestier G, Weber J, Idoumghar L, Muller P (2019) Deep learning for time series classification: a review. Data Min Knowl Discov 33(33):91–963
Fourcade A, Khonsari R (2019) Deep learning in medical image analysis: a third eye for doctors. J Stomatol Oral Maxillofac Surg 120(4):279–288
Gráfová L, Kašparová M, Kakawand S, Procházka A, Dostálová T (2013) Study of edge detection task in dental panoramic X-ray images. Dentomaxillofac Radiol 42:20120,391:1-20120,391:12
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the 2016 IEEE conference on computer vision and pattern recognition. IEEE, pp 770–778
Hwang JJ, Jung YH, Cho BH, Heo MS (2019) An overview of deep learning in the field of dentistry. Imaging Sci Dent 49(1):1–7
Jayachandran S, Aruna P, Preethi M, Yuvaraj M (2019) Ascertaining of age by Raman spectroscopic analysis of apical dentin–a forensic study. J Forensic Dent Sci 11(1):11–15
Jones H (2018) Deep Learning. Createspace Independent Publishing Platform, Scotts Valley
Kašparová M, Halamová S, Dostálová T, Procházka A (2018) Intra-oral 3D scanning for the digital evaluation of dental arch parameters. Appl Sci 8:1838:1-1838:9
Kingma D, Ba J (2015) Adam: a method for stochastic optimization, pp 1–15. CoRR ArXiv:1412.6980
Lamont R, Egland P (2015) Chapter 52: Dental caries. In: Tang YW, Sussman M, Liu D, Poxton I, Schwartzman J (eds) Molecular medical microbiology, 2nd edn. Academic Press, London, pp 945–955
Lee J, Kima D, Jeonga S, Choi S (2018) Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent 77:106–111
Li Z, Wang S, Fan R, Cao G, Zhang Y, Guo T (2019) Teeth category classification via seven-layer deep convolutional neural network with max pooling and global average pooling. Int J Imaging Syst Technol 29:577–583
Mandic D, Chambers J (2001) Recurrent neural networks for prediction. Wiley, Chichester
Mansour R, Al-Marghilnai A, Hagas Z (2019) Use of artificial intelligence techniques to determine dental caries: a systematic review. Int J Neural Netw Adv Appl 6:1–11
Mirza B, Wang W, Wang J, Choi H, Chung N, Ping P (2019) Machine learning and integrative analysis of biomedical big data. MDPI Genes 10:87:1-87:29
Mu C, Li G (2019) Research progress in medical imaging based on deep learning of neural network. Chin J Stomatol 54(7):492–497
Ng C, Almaz E, Simon J, Fried D, Darling C (2019) Near-infrared imaging of demineralization on the occlusal surfaces of teeth without the interference of stains. J Biomed Opt 24(3):036,0021-036,0028
Nishizaki H, Makino K (2019) Signal classification using deep learning. In: Proceedings of the 2019 IEEE international conference on sensors and nanotechnology. IEEE, Penang, Malaysia, pp 1–4
Oh S, Hagiwara Y, Raghavendra U, Rajamanickam Y, Arunkumar N, Murugappan M, Acharya U (2018) A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput Appl 33:1–16
Ordonez F, Roggen D (2016) Deep convolutional and LSTM recurrent neural networks for multimodal wearable activity recognition. MDPI Sens 16:115:1-115:25
Pitts N, Zero D, Marsh P, Ekstrand K, Weintraub J, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A (2017) International caries detection and assessment system (ICDAS): a new concept. Nat Rev Dis Primers 3:17,030:1-17,030:16
Prashar P (2014) Neural networks in machine learning. Int J Comput Appl Technol 105(14):1–3
Procházka A, Charvátová H, Vyšata O, Jarchi D, Sanei S (2021) Discrimination of cycling patterns using accelerometric data and deep learning techniques. Neural Comput Appl 33:7603–7613
Procházka A, Dostál O, Cejnar P, Mohamed H, Pavelek Z, Vališ M, Vyšata O (2021) Deep learning for accelerometric data assessment and ataxic gait monitoring. IEEE Trans Neural Syst Rehabil Eng 29:33434,133:1-33434,133:18
Procházka A, Dostálová T, Kašparová M, Vyšata O, Charvátová H, Sanei S, Mařík V (2019) Augmented reality implementations in stomatology. MDPI Appl Sci 9:2929:1-2929:13
Procházka A, Kuchyňka J, Vyšata O, Cejnar P, Vališ M, Mařík V (2018) Multi-class sleep stage analysis and adaptive pattern recognition. Appl Sci 8(5):697:1-697:14
Procházka A, Kuchyňka J, Vyšata O, Schatz M, Yadollahi M, Sanei S, Vališ M (2018) Sleep scoring using polysomnography data features. Signal Image Video Process 12(6):1043–1051
Procházka A, Vyšata O, Mařík V (2021) Integrating the role of computational intelligence and digital signal processing in education. IEEE Signal Process Mag 38(3):154–162
Procházka A, Vyšata O, Vališ M, Tupa O, Schatz M, Mařík V (2015) Bayesian classification and analysis of gait disorders using image and depth sensors of Microsoft Kinect. Digit Signal Process 47(12):169–177
Raza A, Mehmood A, Ullah S, Ahmad M, Choi G, On B (2019) Heartbeat sound signal classification using deep learning. MDPI Sens 19:48191–481915
Sangaiah A (2019) Deep learning and parallel computing environment for bioengineering systems. Elsevier, Amsterdam
Schwarz R, Gao W, Daye D, Williams M, Richards-Kortum R, Gillenwater A (2008) Autofluorescence and diffuse reflectance spectroscopy of oral epithelial tissue using a depth-sensitive fiber-optic probe. Appl Opt 47(6):825–834
Schwendicke F, Golla T, Dreher M, Krois J (2019) Convolutional neural networks for dental image diagnostics: a scoping review. J Dent 91:103,226
Yang J, Xie Y, Liu L, Xia B, Cao Z, Guo C (2018) Automated dental image analysis by deep learning on small dataset. In: Proceedings of the 42nd IEEE international conference on computer software & applications. IEEE, Tokyo, Japan, pp 492–497
You W, Hao A, Li S, Wang Y, Xia B (2020) Deep learning-based dental plaque detection on primary teeth: a comparison with clinical assessments. BMC Oral Health 20(141):1–7
Zhang Z, Zhang B, Akiduki T (2019) Specular reflection surface defects detection by using deep learning. In: Proceedings of the 3rd international conference on information system and data mining. ACM Press, Houston, USA, pp 6–10
Acknowledgements
This research was supported by the project GA UK No. 52220 of the Charles University. No ethical approval was required for this study.
Author information
Authors and Affiliations
Contributions
AP was responsible for the mathematical methods of data processing, JC recorded all data, OV interpreted results, and DM contributed to data preprocessing and to evaluation of results. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Procházka, A., Charvát, J., Vyšata, O. et al. Incremental deep learning for reflectivity data recognition in stomatology. Neural Comput & Applic 34, 7081–7089 (2022). https://doi.org/10.1007/s00521-021-06842-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06842-6