Abstract
Epilepsy remains one of the most common chronic neurological disorders; hence, there is a need to further investigate various models for automatic detection of seizure activity. An effective detection model can be achieved by minimizing the complexity of the model in terms of trainable parameters while still maintaining high accuracy. One way to achieve this is to select the minimum possible number of features. In this paper, we propose a long short-term memory (LSTM) network for the classification of epileptic EEG signals. Discrete wavelet transform (DWT) is employed to remove noise and extract 20 eigenvalue features. The optimal features were then identified using correlation and P value analysis. The proposed method significantly reduces the number of trainable LSTM parameters required to attain high accuracy. Finally, our model outperforms other proposed frameworks, including popular classifiers such as logistic regression (LR), support vector machine (SVM), K-nearest neighbor (K-NN) and decision tree (DT).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Epilepsy is one of the most common chronic neurological diseases; it is characterized by abnormal electrical activity in the brain [1]. The symptoms include involuntary movements (seizures), loss of consciousness and loss of control of bowel and/or bladder function. According to the World Health Organization (WHO), approximately 50 million people currently suffer from epilepsy [2].
A seizure is defined as a transient period of excessive or synchronous neuronal activity. Seizures can be caused by a variety of conditions. Epilepsy is one such condition and is defined by the number and frequency of unprovoked seizures [3]. Electroencephalogram (EEG) is a valuable clinical tool for the diagnosis and treatment of epilepsy as it allows for measurement of the brain’s electrical activity. EEG signals are complex, non-stationary, nonlinear and random [4]. This is due to the inherently complex nature of a system made up of billions of interconnected neurons. There are three major steps to analyze EEG signals, namely preprocessing, feature extraction and classification. The preprocessing stage focuses on artifact removal, while the feature extraction stage aims to identify the features of the signal that represent epilepsy. The classification stage employs techniques that classify the signal based on the extracted features.
Classification is an important part of signal analysis. Several classifiers, such as support vector machine (SVM), K-nearest neighbor (K-NN) and artificial neural network (ANN), have been proposed for epilepsy classification. Li et al. [5] proposed a novel method of classifying normal, interictal and epileptic signals using a wavelet-based analysis (EA) neural network ensemble (NNE). Bhattacharyya et al. [6] used a tunable-Q wavelet transform-based multiscale entropy measure for automated classification of epileptic EEG signals. After decomposing with the tunable-Q wavelet transform (TQWT) into subbands, K-nearest neighbor (K-NN) entropies from various subbands were cumulatively estimated and fed into a support vector machine (SVM) classifier using the wrapper-based feature selection method. A satisfactory result was obtained. Misiūnas et al. [7] proposed an EEG classification algorithm based on the type of epilepsy: benign focal childhood epilepsy and structural focal epilepsy. Basically, the algorithm estimates the spike parameter of the signal and utilizes an ANN for type classification. These researchers have used large data samples for proof of concept. Another approach to classification involves utilization of epileptic EEG energy. In this vein, Fasil and Rajesh [8] used the time-domain exponential energy of EEG for classification. However, the performance of the algorithm varied greatly when applied to two different datasets. Efforts have also been made to classify partial epilepsy [9]. In this regard, a “Wave_clus spike” sorting algorithm was applied to patients’ simultaneous electroencephalogram–functional magnetic resonance imagining (EEG–fMRI) records in order to classify interictal epileptic form discharges (IED). An attempt has also been made to identify the pre-seizure state on EEG records [10]. In this novel seizure prediction method, ten sub-frequency EEG datasets were generated from the initially recorded signals. After the generated sub-frequency was used to mine the EEG features, a generalized regression neural network (GRNN) was applied to separate the EEG signal clips into normal and pre-seizure files. In addition, an automatic EEG-based epileptic seizure detection system using fragmented feature extraction and a least-square support vector machine (LV-SVM) classifier has been proposed [11]. In that system, EEG preprocessing is accomplished with a Butterworth bandpass filter and statistical features are extracted from the EEG signal fragments. A mean accuracy of 97.19% was obtained.
LSTM is another popular deep learning technique that has recently been applied to classification. Ahmedt-Aristizabal et al. [12] proposed a deep learning classification system based on LSTM for epileptic signals. The proposed system eliminates the need for preprocessing and laborious feature extraction techniques. The system provided an average testing accuracy of 91.25% with two different configurations, both of which require a very large number of trainable parameters. The first (one-to-one) configuration requires about 17,000 trainable parameters, while the second (many-to-one) requires about 116,033 trainable parameters. Hussein et al. proposed a robust epilepsy detection system that is capable of handling noisy EEG data using LSTM. A promising result was recorded for noisy data [13]. However, the proposed LSTM network needed three layers to achieve good accuracy. No information about the number of neurons was given. In another study, a deep learning approach to feature learning from EEG data that attempted to extract the correlation between data samples was proposed [14]. The architecture recorded high scores with different numbers of classes, from two to five classes. However, about 30,610 trainable parameters are needed to learn the dependencies between the EEG signals across the same classes. The LSTM network converges after about 2400 iterations with 40 epochs. Thus, it required a long training time. However, in 2019, a deep network for robust detection was optimized in another work [15]. The optimized network reduced the number of LSTM units and iterations to 80 and 2000, respectively. Bidirectional LSTM (Bi-LSTM) is another popular version of LSTM. Abdelhameed et al. [16] proposed a Bi-LSTM network for EEG detection. CNN was employed for preprocessing and feature extraction of the EEG data. The authors conducted experiments using 2 and 3 classification problems. However, unlike our approach, which groups preictal and interictal data in a healthy subject, the two-class problem only considers preictal and ictal data. Furthermore, Hu and Yuan [17] proposed an identification system for epileptic EEG signals using a deep Bi-LSTM network. Linear features were extracted after the signal was subjected to a bandpass filter. The linear features include mean, standard, skewness and variance. The proposed method recorded an accuracy of 98.56%.
In this paper, we propose an LSTM for the classification of epileptic EEG signals using an optimal wavelet feature selection algorithm based on correlation and P-value. Discrete wavelet transform (DWT) is employed to remove noise and extract 20 eigenvalues as features. Optimal features were then selected using correlation and P-value analysis. Our proposed method resulted in a significant reduction in the number of trainable LSTM parameters needed to attain high accuracy.
The rest of the paper is organized as follows. In Sect. 2, we present our proposed framework and the algorithm used for feature extraction and selection. Section 3 discusses the experimental results of the LSTM classification model with various features. Finally, Sect. 4 concludes the paper.
2 Materials and methods
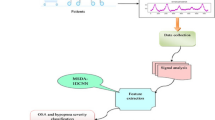
The proposed framework consists of four major steps: EEG data collection, feature extraction, feature selection and classification. Raw EEG data are obtained from patients and normal subjects and then subjected to DWT to remove noise and extract 20 eigenvalues as features. From the extracted features, the optimal features that effectively provide enough information to discriminate between normal and patient EEG records are then selected. Finally, an LSTM classifier is built to classify the EEG based on the selected features. Various experiments were conducted on various architectures and structures using the classifier. Figure 1 shows the proposed framework.
2.1 EEG data collection
The data used in this study are publicly available from the Epilepsy Research Center at Bonn University in Germany [18]. The EEG dataset consists of five subsets, denoted A–E, and each subset contains 100 single-channel EEG segments of duration 23.6 s taken at a sampling rate of 173.61 Hz. Each segment consists of 4097 sampling points. A and B contain EEG data on five healthy subjects while the subjects blinked their eyes and kept their eyes closed, respectively. C and D contain epileptic EEG signals collected from the hippocampal formation and epileptogenic zone, respectively. For C and D, the data were measured during seizure-free intervals, while E was measured during seizure episodes. Sets A–D are considered normal EEG datasets, while E is classified as an epileptic dataset. Each category has 100 samples. To establish the model system, the data samples were randomly split into training and testing datasets containing 80 and 20% of the data, respectively. Table 1 shows the composition of the dataset.
2.2 Methods
An LSTM modeling flowchart is shown in Fig. 2. The data are split into training–validation data and testing data in a 4:1 ratio. Furthermore, the training–validation data are grouped into training and validation data in a 3:1 ratio. Validation is conducted after each training epoch, while testing is conducted with the final model after validation. As indicated in the diagram, stochastic mini-batches are used to train the model. This approach divides the training data into mini-batches, calculates the model error of each batch and then updates the model weight accordingly. This way, less dedicated GPU memory is required to design a model with a higher learning rate; this approach also helps to avoid the training process becoming trapped in local minima of the loss function [19].
2.2.1 Wavelet transform of EEG data
A wavelet is a wave-like vibration accompanied by an amplitude that repeats, increases and decreases around zero. Wavelets have useful features for signal processing; for example, they can be used to extract information from an unknown signal in combination with a known signal through a convolution technique. This technique is often used to extract useful information from audio signals or images as well as from a variety of other types of data. Wavelets are also useful for wavelet-based decompression algorithms designed to minimize loss and restore raw information [20].
Wavelet transform can be thought of as a time–frequency representation of unique signals that are used to model signals, systems and processes through a combination of wavelet functions. A wavelet transform is expressed as an arbitrary waveform after being scaled (for scaling, one small waveform is used as a pattern to be transited, enlarged, or reduced).
A low-pass filter L which satisfies the standard quadrature mirror filter condition can be used to specify all wavelet transforms:
where the filter’s z-transform is denoted by \(L\left( z \right)\). On the other hand, the high-pass filter H is defined as
Hence, a sequence of filters can be expressed as:
Given the initial condition \(L_{0} \left( z \right) = 1\), the sequence of filters can be expressed by a two-scale relation in the time domain:
where \(\left[ \right]_{ \uparrow r}\) denotes up-sampling by a factor of r and k is the discrete-time sampled. Therefore, the normalized wavelet \(\varphi_{i,l} \left( k \right)\) and scale basis function \(\psi_{i,l} \left( k \right)\) can be defined as
where \(2^{i/2}\) denotes the inner product normalization, i is the scale parameter and l is the translation parameter. The DWT decomposition can be expressed as
where \({\mathcal{H}}_{\left( i \right)} \left( l \right)\) is the detail coefficient and \({\mathcal{L}}_{\left( i \right)} \left( l \right)\) is the approximation coefficient at resolution i [21].
In this work, we apply DWT to each EEG channel for feature extraction. DWT is particularly suitable for representing non-stationary signals, such as EEG signals, because it provides optimal resolution in both time and frequency [22].
2.2.2 Feature extraction
In order to extract the features, each EEG signal, X(z), is decomposed at different frequency bands into a detailed and approximate coefficient using DWT. The decomposition is performed by repeatedly dividing the EEG signal into two bands through a high-pass filter and a low-pass filter until the desired level is attained. The detailed coefficients are obtained by filtering the signal through a high-pass filter (H(z)), while the approximations are computed using a low-pass filter (L(z)). The procedure for decomposition of each signal X(z) is shown in Fig. 3.
As the analysis of EEG signals using DWT depends on the number of decompositions and the chosen wavelet function, we adopt four-level decomposition and a Daubechies-2 wavelet function in this work. The Daubechies-2 wavelet function is suitable for EEG because EEG is similar to spike-wave in pattern, and the Daubechies sequence has better scalability and flexibility for weighting boundary problems [1]. On the other hand, four-level decomposition is adopted because it captures the dominant frequency bands in EEG. Table 2 shows the wavelet decomposition level and the corresponding frequency according to EEG frequency.
Twenty features are extracted from the computed detail and approximation coefficients that were obtained by applying the DWT to all EEG channels. Specifically, using higher-order statistics, four different types of nonlinear features were extracted from the obtained detail and approximation coefficients (D1; D2; D3; D4; A4) of each channel. The extracted features in each subband include maximum, minimum, mean and standard deviation coefficients. Thus, 20 features were extracted in total.
The mean represents the average wavelet coefficient of each decomposition level. The mean is obtained using Eq. (7):
where \(D_{L}\) is the number of coefficients in a decomposition level and \(X_{i}\) represents the value of the coefficients i in a decomposition level.
The standard deviation feature is a measure of the variation of the coefficients from the mean values in a decomposition level. It is computed using Eq. (8).
The minimum feature is the minimum value of a coefficient in a specific decomposition level. It is computed using Eq. (9).
where \(X_{L}\) is a group of detail or approximation coefficients under consideration.
The maximum feature is the maximum value of a coefficient at a particular decomposition level. It is computed using Eq. (10).
where \(X_{L}\) is the group of detail or approximation coefficients under consideration.
2.2.3 Feature selection
Reducing the number of features is important in addressing the problem of complexity. Thus, in this section, we present two ways to reduce the number of features—feature selection and feature dimensionality reduction, using correlation coefficient and P-value analysis (CCP) and principal component analysis (PCA).
2.2.3.1 Correlation coefficient and P-value feature (CCP) selection
Correlation coefficients and P-values are the most commonly used statistical tools for establishing a relationship between variables. While a correlation coefficient is used to test for any kind of relationship between variables, the P-value informs us whether or not the result of an experiment is statistically significant [23].
The correlation coefficient is a measure that describes the strength of the relationship between two variables. There are several types of correlation coefficient, including Pearson, Kendall and Spearman coefficients. In this work, we employ the most common type, Pearson correlation. A Pearson coefficient is a number between − 1 and 1, with 1 representing the strongest possible positive correlation and − 1 being the strongest possible negative correlation. Pearson’s formula \(\left( {r_{xy} } \right)\) for a dataset consisting of n pairs of data points is given as:
where n is the sample size, \(x_{i, } y_{i }\) is one pair of data points and \(\bar{x}\) and \(\bar{y}\) are the sample means.
Scientists use statistical analysis to understand, explain and predict natural phenomena [24]. For over 75 years, statistical analysis as a whole has been characterized by using null hypothesis significance testing (NHST). The null hypothesis simply states that there is no relationship between the two features or groups that are being compared. In NHST, the P-value is used as a tool to measure the degree to which the data conflict with the null hypothesis. In other words, given that the null hypothesis is true, the P-value is the probability of obtaining results that are as extreme or more extreme than the actual data. The P-value assesses significance by means of an alpha level which gives the probability of rejecting the null hypothesis when the null hypothesis is true. The CCP method has shown a remarkable improvement in classification accuracy for breast cancer diagnosis [23].
In this work, the null hypothesis is that “the selected combination of dependent variables (features) have no relationship with the independent variable (healthy or patient subject group).” The alpha value is set to 0.05, i.e., p < 0.05. Thus, if the null hypothesis is true, the probability of obtaining a test statistic as extreme or more extreme than the one observed is less than 5%. As illustrated in Algorithm 1, for features with a high correlation of 0.9 and above, one of the two correlated features is selected before P-value analysis is conducted.

2.2.3.2 Principal component analysis (PCA)
Principal component analysis (PCA) is an unsupervised learning algorithm that is typically employed to find patterns in high-dimensional data. Its main goal is to perform data dimensionality reduction, lossy data compression, feature extraction and data visualization. It is also called the Karhunen–Loève transform and is defined as an orthogonal projection of data in low-dimensional linear space. Using PCA, n-dimensional features can be mapped onto a completely new orthogonal projection in k-dimensional space (k < n) [1].
PCA is employed in our work to reduce the dimension of the EEG data. The n-dimensional EEG data (D sample), which have 20 dimensions, are reduced to n′-dimensional data. In this process, the D sample is normalized, and the covariance matrix is computed and subjected to eigenvalue decomposition. n′ eigenvalues corresponding to the largest eigenvalues are then removed and normalized to form the eigenvector matrix. Finally, the sample is transformed to obtain the desired output sample with the required number of features, which is then used as an input to the classifier. In this work, we experimentally determined the optimal number of features that gave the best generalization with the fewest possible trainable parameters.
2.2.4 Classification using long short-term memory (LSTM)
Deep learning is essentially a machine learning method that utilizes many hidden layers to carry out multi-level nonlinear operations in neural networks. With several transformations and multiple hidden neurons, complex functions can be learned to discriminate the response classes in a classification problem. A recurrent neural network is a deep learning tool that was developed for processing of sequential data. It consists of feed-forward neural networks with cyclic connections. It maps the entire history of input in the network to predict each output by leveraging the temporal relationships between the data at each point in time.
The architecture of a simple RNN is shown in Fig. 4. In the diagram, each node represents a layer of the network at one point in time. The network nodes consist of a weighted connection between the input and hidden layer, hidden and hidden layer and hidden and output layer. The weighted connections from input to hidden layer, hidden to hidden layer and hidden to output layer are represented by the matrices U, W and V, respectively. The final weight matrix is passed through a softmax function to produce a scalar Y value, which is then classified as a binary variable, Ŷ. The loss function is then applied to compare Y-actual and Y-predicted (Ŷ). However, the current RNN has a vanishing gradient problem in which a given input influences the hidden layer and, subsequently, network output [25]. This either decays exponentially or explodes exponentially over time as the data go through transformations on the network. Two popular solutions to this issue have been developed: long short-term memory (LSTM) and gated recurrent unit (GRU).
The current research deploys LSTM architecture. It is made up of connected subnetworks, collectively called a memory block, which remembers inputs for a long time. Each block contains at least one self-connected accumulator cell and several multiplicative units, such as the input gate, forget gate and output gate. Information is stored and accessed through the gates by assigning a counter, such as 0 and 1. Figure 5 presents the standard RNN and the LSTM employed in this research. LSTM is suitable for exploring EEG data due to its ability to explore the temporal correlation between EEG signals [26].
The proposed framework was modeled and validated using the Python programming language (version 3.6). The model is implemented using TensorFlow v1.0 at the backend and Keras API at the front end.
2.2.5 Performance evaluation metric
The performance of the proposed model was evaluated at various stages with feature extraction using DWT, feature selection using CCP and PCA. The performance evaluation metrics include accuracy, precision, specificity, sensitivity and \(F_{1}\) and are defined as follows:
where TP, TN, FP and FN are the number of true-positive cases, true-negative cases, false-positive cases and false-negative cases, respectively.
3 Results and discussion
This section presents the results of the methods employed for feature extraction and selection described in Sect. 2.2. Likewise, the process of obtaining optimal hyperparameter settings for LSTM training, validation and testing is presented. Then, the performance of the LSTM model is compared against that of logistic regression, support vector machine, K-nearest neighbor and random forest methods, GRU, bidirectional LSTM (BiLSTM) and other algorithms proposed by other authors.
3.1 Feature extraction
In this section, we examine the nonlinear features extracted from all of the EEG channels by applying a wavelet function (described in Sect. 2.2.1). The wavelet analysis decomposes the EEG signals into separate subband channels to unravel the superposition of different structures. As presented in Table 2, the EEG signal is decomposed into four levels for extraction of the dominant frequency component. The wavelet coefficients at various levels are retained to provide the frequencies required for signal classification. For EEG signals, the Daubechies wavelet of order 2 (db2) is the most efficient at detecting various changes [21]. Thus, db2 was applied to compute the wavelet coefficient of the details, D1–D4, and the approximation, A4.
Meanwhile, normal and abnormal brain waves can be distinguished via frequency analysis. As shown in Fig. 6, a 3-second continuous signal from a healthy brain wave differs from that of a brain wave during a seizure. The difference stems from pathological changes in brain tissue caused by the seizure [27].
A rectangular window was applied to each segment of the EEG signal to extract features such that 256 discrete data points were created while the signal remained stationary in that interval. For each segment of the EEG, the detail coefficients (Dk, k = 1, 2, 3, 4) at the four levels were 129, 66, 34 and 18, respectively. The approximation coefficient (A4) at the fourth level was 18. Therefore, a total of 265 wavelet coefficients were extracted for each segment. These obtained coefficients are a compact representation of the EEG signal distribution in both time and frequency. Finally, 20 statistical features were extracted in order to reduce the dimensionality of feature vectors. The statistical features obtained for each of the four detail coefficients and the approximation coefficient include maximum, minimum, mean and standard deviation.
Figure 7 shows the boxplot of each subband. In the figure, maximum, minimum and standard deviation can be used to discriminate between normal/healthy and patient EEG signals. The EEG of the normal subject has a lower mean and distribution compared to the epileptic EEG in the maximum and standard deviation features, while the epileptic EEG has a lower mean and broader distribution in the minimum feature. The mean feature, on the other hand, only significantly differs between D4 and A4. Furthermore, the EEG of the normal subject has more outliners than the epileptic EEG.
In this work, we maintain the outliers and focus on selecting the optimal features and building an effective model that can discriminate between the two signals. Thus, we further compute the coefficient matrix to understand the relationship between the features.
3.2 Dimensionality reduction analysis
As shown in the coefficient matrix of the features in Fig. 8, some of the features are strongly correlated. This implies that the features that are strongly correlated with the dependent variable, i.e., classes, are linearly dependent and are therefore correlated with one another. Thus, we investigated two methods of reducing the dimensionality of the data, CCP and PCA.
3.2.1 CCP analysis
In this method, we compute and compare the correlation between features and eliminate one each pair of features that have a correlation coefficient greater than or equal to 0.9. This reduces redundancy of features that have the same effect on the training of the model. Figure 9 shows the correlation matrix after the elimination.
After the elimination of similar features and the formation of a new correlation matrix, features were selected based on P-value. As discussed in Sect. 2.2.3, the significance level (alpha value) was set to 0.05, which indicates that the probability of attaining equal or more extreme results assuming that the null hypothesis is true is 5%. Thus, the null hypothesis, “The selected combination of dependent variables (features) has no relationship with the independent variable (healthy or patient subject group)” is rejected when the P-value is less than the specified alpha value (level of significance).
The process of forming the reduced set of features (X) is illustrated in Fig. 10. In the initial stage, the feature subset (X) contained NULL; we then employed backward elimination to eliminate all the feature pairs that have a P-value lower than alpha in each iteration until all of the features were evaluated. In the end, three features were selected and added to the subset (X). The selected features are D1 minimum, D4 mean and A4 minimum.
Figure 11 presents the distribution of the selected features. The D4 mean feature is normally distributed, while D1 minimum and A4 minimum features are negatively skewed.
The computational complexity of the proposed CCP selection algorithm is given as follows:
-
Time complexity
Computing inter-feature correlation between n features: \(O\left( {n^{2} } \right)\).
Finding and removing one of each pair of features with a correlation coefficient of 0.9 or greater: \(O\left( n \right)\).
Computing the P-value:\(O\left( n \right)\).
Time taken to compare P-value with significance level: \(O\left( n \right)\).
-
Space complexity
Space for storing n number of features: \(O\left( n \right)\)
Given the total time complexity of the CCP, the time complexity is quadratic; thus, the time of execution increases for a large number of features. Also, the space complexity increases linearly with an increase in n, i.e., number of features stored in memory during each run.
3.2.2 PCA
In this subsection, we present the results of feature dimensionality reduction using PCA. Based on the correlation matrix in Fig. 8, nine or fewer features may be sufficient to classify a signal into normal/healthy and patient groups. Thus, we experimented with the use of 9, 6 and 3 features to observe the performance of the model with a reduced number of features. The plot of the coefficient matrix is given in Fig. 12.
3.3 Classification performance
In this section, the result of various training, validation and testing procedures conducted to identify the optimal LSTM model is presented. Then, the performance of the LSTM model compared to logistic regression, SVM, K-NN, GRU, BiLSTM and other algorithms is presented.
3.3.1 Tuning the hyperparameters and architectural components of the LSTM model
The first part of our classification experiment aims to determine the hyperparameters and LSTM model structure that give the best generalization performance. Table 3 presents the LSTM hyperparameter under investigation. The performance of various configurations based on depth, number of neurons, optimizer and accuracy is presented in the subsequent subsection.
However, before we present the performance of various configurations, a default architecture consisting of three layers (128–256–256 neurons in the three layers) was used to determine the dropout, epoch and batch size. A dropout is placed between each layer. Then, the softmax function and Adam optimization algorithm are used as an output activation function and optimizer, respectively. The dataset with 20 features extracted using DWT, as discussed in Sect. 3.1, is used in this experiment. The detailed hyperparameter settings for our network are illustrated in Fig. 13.
Dropout is a regularization technique used to minimize overfitting. Basically, several units are randomly dropped from the network based on the affine transformation. Thus, we first experimented to determine the optimal dropout for epilepsy classification.
The performance of each optimizer was investigated by varying the dropout in a range of 0.1 to 3.0 and an interval of 0.05. As indicated in Table 4, Adam and RMSprop had the best generalization of 97% accuracy at 0.25 dropout. SGD gave the worst result, 80% accuracy across the entire dropout range. Figure 14 shows a histogram of the results. Based on this performance analysis, a dropout of 0.25 was selected for the model.
Based on the default configuration, we conducted ten experiments to determine the optimal batch size for training and validation using 5 epochs. We considered batch sizes of 1, 2, 3, 4 and 5. Figure 15 presents the boxplot of the ten experiments for each batch. The model performs poorly with a batch size of 1, with a mean accuracy of less than 0.97, minimum accuracy of 0.95 and maximum accuracy of 0.97. Batch sizes of 3 and 4 resulted in the best performance, with a mean accuracy of 0.97, minimum accuracy of 0.96 and maximum accuracy of 0.98. However, considering speed, a batch size of 4 was chosen for the model.
We also investigated the influence of epoch on accuracy. We used the optimal batch size of 4 and dropout of 0.25 to conduct ten experiments on each epoch (10, 15, 20 and 25). As indicated in Fig. 16, an epoch of 10 was associated with poor performance, as the accuracy ranged widely from 0.95 to 0.96. On the other hand, an epoch of 25 gave the best performance as it maintained an accuracy of 0.96, though there is 1 outliner with an accuracy of 0.95.
3.3.2 Classification with wavelet-extracted features
In this section, we present the model performance using the 20 extracted wavelet features. We also investigate the accuracy of the various optimizers listed in Table 3. We trained the model using the default architecture discussed in 3.3.1, with batch size, dropout and epochs of 4, 0.25 and 25, respectively.
After conducting several experiments with various configurations, we determined that a three-layer network with 128-256-256 neurons gave the optimal generalization. Figure 17a presents the training performance of various optimizers, with Adam having the best training performance of about 0.97. Generally, except for SGD, the performance of all optimizers progressively improved during the training despite some setbacks.
Likewise, we observed progressive improvements in the validation performance of the optimizers for Adam and RMSprop, as shown in Fig. 17b. However, AdaGrad showed instability and varied in accuracy between 0.92 and 0.98, while SGD failed to learn and maintained an accuracy of just 0.84 across the training epoch.
Based on the training and validation performances, Adam was chosen as the best optimizer for our model. The model was then tested with the testing data, and an accuracy of 0.99 was recorded. The resulting confusion matrix is presented in Fig. 18.
3.3.3 Classification with dimension reduction
Though we obtained high accuracy using 20 features, there was still a need to reduce the complexity of the LSTM network by reducing the depth of the layers and the number of neurons in each layer. CCP and PCA feature selection methods were employed to achieve that aim. Various configurations were investigated to obtain the best model with the highest classification accuracy.
3.3.3.1 CCP feature selection classification performance
The optimal features selected in Sect. 3.2.1 are used in this experiment. We conducted several experiments with two different layer structures while varying the numbers of neurons, as indicated in Table 3. With a three-layer architecture with 128-256-256 neurons, Adam performed the best, with an overall accuracy of 0.92. In the training session shown in Fig. 19a, the performance of all optimizers except SGD progressively improved. Adam attained the highest training accuracy of about 0.95 before settling slightly lower. During validation, RMSprop and Adam start with an accuracy above 0.86. Adam finally settles at 0.94, while RMSprop experienced overfitting and settled back to the starting point after attaining an accuracy of about 0.95 during the session. Conversely, the performance of AdaGrad oscillates between 0.93 and 0.95. Figure 19b presents the validation performance.
In the experiment with a two-layer architecture, the best performance was obtained with RMSprop and a 16–32 neuronal configuration. In the training session, with the exception of SGD, the performance of all of the optimizers progressively improved, as shown in Fig. 20a. The learning performance of Adam was fastest, followed by RMSprop and then AdaGrad. RMSprop and Adam attained high performance of more than 0.92, while SGD failed to learn and exhibited an accuracy of 0.79 as training progressed. The ranking of optimizers during validation was similar to that during training, as presented in Fig. 20b. However, Adam and RMSprop attained a peak performance of about 0.93 with about 13 epochs. Beyond 13 epochs, the performance of both oscillated between 0.92 and 0.93.
Comparing the test performances of the two configurations, i.e., three- and two-layer, discussed above, showed that the two-layer architecture with the RMSprop optimizer and 16–32 neurons performed the best, with an overall accuracy of 0.99. The three-layer configuration showed the second best performance, with an overall accuracy of 0.92 with the Adam optimizer and 128-256-256 configuration. Table 5 presents the model performance evaluation of the best configuration for CCP.
Figure 21 shows the model performance evaluation based on precision, recall and F1 score. Except for recall for Patient data in the three-layer configuration, which had the worst performance of 0.58, all others showed a perfect recall score of 1. In terms of precision, both the two-layer and three-layer configurations showed a perfect score of 1 for both healthy and epileptic subjects, while healthy and patient subjects for three layers and two layers scored 0.90 and 0.95, respectively. In addition, three-layer and two-layer architecture showed F1 scores of 0.95 and 1, respectively, for healthy subjects, while the F1 scores for patients were 0.73 and 0.90, respectively. Based on these findings, a two-layer configuration with 16–32 neurons and RMSprop is the optimal model. The low performance of the three-layer configuration is due to overfitting: As there are more trainable parameters that have adapted to the training data, model performance is poor on a new set of data. As presented in the confusion matrix for the two-layer model (Fig. 22), just one healthy subject was misclassified as a Patient.
3.3.3.2 Classification with PCA reduction
PCA is the second technique employed to reduce the dimensionality of the data and thus reduce the complexity of the model. Three sets with different numbers of features were selected and experimented upon. The number of features selected includes 3, 6 and 9. The classifier configuration considered for the experiment is the same as that described in Sect. 3.3.1.
The training performance of all of the optimizers progressively improved for the three feature sets, with the exception of SGD. Specifically, 3 features with a two-layer configuration (16–32 neurons) attained about 0.95 accuracy, while 6 and 9 features with a two-layer configuration (64–128 neurons) both reached about 0.97 accuracy. Figure 24a–c presents the training performances of the best models.
In the validation experiment, use of 6 and 9 features led to overfitting; the accuracy in those cases was about 0.92 and 0.96, respectively. On the other hand, 3 features did not cause overfitting, and the validation accuracy of the model was about 0.97 accuracy for all optimizers except SGD. Thus, as shown in Fig. 24d–f, the 3-feature model is sufficient for the classification problem.
We next evaluate the performance of the models with testing data. As shown in Table 6, the models with 9 and 6 features had an overall accuracy of 0.98 with two-layer (64–128 neurons, AdaGrad) and two-layer (64–128 neurons, RMSprop) architecture, respectively. On the other hand, the 3-feature model had the best generalization of 0.99 with two-layer architecture, 16–32 neurons and the AdaGrad optimizer. Furthermore, the 3-feature model requires the lowest number of neurons to attain high classification accuracy.
Figure 23 shows the model performance evaluation based on precision, recall and F1 score. The 3-feature model exhibited a perfect score of 1 in precision and recall for healthy and patient subjects, respectively, while it scored about 0.94 and 0.97 in precision and F1 score, respectively, for Patient subjects. For the 6-feature model, scores of 0.99 and 0.93 were recorded for healthy and patient subjects, respectively, in precision, recall and F1-score. Scores of 0.99 and 0.94 were recorded for healthy and patient subjects, respectively, in precision, recall and F1-score in the 9-feature model.
Based on the training, validation and testing performance, the 3-feature model with two-layer architecture, 16–32 neurons and the AdaGrad optimizer, is the optimal model developed with the PCA method of dimensionality reduction. We also observed that when we increased the depth of the network to three layers with 16–32–32 neurons, RMSprop and Adam attained equal accuracy scores of 0.99. However, the two-layer configuration is better as it is less complex (has fewer trainable parameters). As presented in the confusion matrix for the two-layer model in Fig. 25, just one healthy subject was misclassified as a Patient.
3.4 Model performance evaluation
3.4.1 Performance evaluation of the LSTM model based on CCP and PCA
Both the CCP and PCA feature selection methods resulted in features that gave high accuracy scores of 0.99. The best CCP-based model was the LSTM two-layer configuration with 16–32 neurons and RMSprop. On the other hand, the best model for PCA dimensionality reduction to 3 features was achieved with LSTM two-layer architecture, 16–32 neurons and AdaGrad. However, considering precision, recall and F1 score, the performance of the models varied in regard to healthy and patient subjects, as shown in Fig. 26. Both CCP and PCA classified healthy subjects with a perfect score of 1.0 for precision, while classification of Patient subjects with CCP and PCA showed precision of 0.95 and 0.94, respectively. Patient subjects were associated with a perfect score of 1.0 for recall in both CCP and PCA, while healthy subjects were associated with a recall score of 0.99 in both CCP and PCA. Except for PCA, where Patient subjects showed an F1 score of 0.97, all others resulted in F1 scores of 0.97. Thus, CCP performed better than PCA, as PCA resulted in a lower performance of 0.94 (precision) and 0.97 (F1 score) in recognizing Patient subjects.
3.4.2 Performance evaluation of the GRU and BiLSTM models based on CCP and PCA
We next conducted several experiments using the selected features with the GRU and BiLSTM models. The optimal hyperparameters were tuned using the same procedures as used for LSTM. The optimal GRU model was obtained with a batch size, number of epochs and dropout of 3, 25 and 0.15, respectively. On the other hand, the best BiLSTM model was attained with a batch size and number of epochs of 4 and 25, respectively. BiLSTM was found to generalize better on this dataset without dropout. In both cases, the training and validation accuracy progressively increased during the training and validation sessions.
In the BiLSTM model, the CCP features selection algorithm resulted in a maximum accuracy of 0.96 with two-layer architecture, 128–256 neurons and Adam, as indicated in Fig. 27. Meanwhile, both PCA and DWT showed a highest accuracy value of 0.98 with 9 and 20 features, respectively. The optimal PCA performance was obtained with three layers, 64–128–128 neurons and Adam, while the optimal DWT model was obtained with three layers, 128–256–256 neurons and AdaGrad.
In terms of the number of trainable parameters, PCA requires the fewest parameters (307,292), followed by CCP (1,204,226) and DWT (2,799,138), as presented in Fig. 28. PCA is thus most applicable to BiLSTM as it gave a high accuracy of 0.98 with fewer features and trainable parameters when compared to DWT.
The GRU model attained a perfect accuracy of 1.0 with 6 and 9 PCA features, followed by DWT and CCP with an accuracy of 0.99 and 0.96, respectively. As indicated in Fig. 29, 6-feature PCA and DWT required three-layer, 64–128–128 neuron configurations while CCP and 9-feature PCA required two layers, 16–32 neurons to attain optimal performance.
6 and 20 features using PCA and DWT required an equal number of trainable parameters (187,266) with RMSprop and Adam, respectively, to attain optimal performance. Meanwhile, 3 and 9 features using CCP and PCA, respectively, attained optimal performance with 6018 trainable parameters and Adam, as shown in Fig. 30. 6- and 9-feature PCA is more suitable for the GRU model as they showed a perfect accuracy of 1.0. However, there is a trade-off between the number of features and trainable parameters: 6 features required more than 31 times the number of trainable parameters compared to 9 features. Therefore, when it is imperative to have fewer features, use of 6 PCA features is the best option. But if the number of trainable parameters is critical, 9 PCA features are best.
Consequently, the CCP feature reduction algorithm is not suitable for the BiLSTM and GRU models as both recorded a maximum accuracy of 0.96, which is low compared to the accuracy of the LSTM model (0.99), as shown in Fig. 31. Although both required two-layer architecture, the number of neurons and trainable parameters varied. Likewise, the best optimizers differed; the best performance of the LSTM, BiLSTM and GRU models was attained with RMSprop, AdaGrad and Adam, respectively.
The LSTM model outperformed the other models in terms of precision, recall and F1 score, as shown in Fig. 32. The LSTM model showed a perfect precision score of 1.0 and a recall and F1 score of 0.99 for healthy subjects, while BiLSTM and GRU demonstrated 0.97, 0.99, 0.98 and 0.96, 0.99, 0.98 precision, recall and F1 score, respectively. Likewise, LSTM outperformed the other models in Patient subjects, as it showed 0.95, 1.00 and 0.98 precision, recall and F1 scores, respectively. Meanwhile, the BiLSTM and GRU models attained 0.92, 0.80, 0.86 and 0.94, 0.85, 0.85 precision, recall and F1 scores, respectively.
3.4.3 Evaluation of the LSTM model based on trainable parameters
PCA, CCP and the 20 features without feature selection or reduction all resulted in an accuracy of 0.99 in the LSTM model. However, given the complexity of the LSTM network, the 20 features extracted by DWT require about 996,354 trainable parameters to attain that high accuracy of 0.99. At the same time, the 3 features that resulted from PCA reduction required 15,938 trainable parameters in LSTM with three-layer architecture. On the contrary, the two-layer LSTM network required about 7618 parameters for training when 3 features were selected using CCP and PCA as shown in Fig. 33. Even though PCA requires the same number of trainable parameters as CCP, as established in Sect. 3.4.1, CCP is the optimal method to reduce complexity. Thus, the use of CCP for feature selection significantly reduced the number of required training parameters by about 13,000%.
We also compared the number of trainable parameters required by our framework vs. other algorithms. As shown in Fig. 34, the algorithm proposed by Hussein et al. [14] requires about 30,610 trainable parameters. Even though the algorithm achieves 100% accuracy, it may not be suitable for lightweight applications such as mobile devices. Also, training requires 2400 iterations with 40 epochs; by comparison, ours requires just 25 epochs to attain 0.99 accuracy. Likewise, the LSTM model proposed by Ahmedt-Aristizabal et al. [12] requires about 116,033 trainable parameters. Even with a significant number of parameters, the accuracy recorded was less than that of our framework.
3.4.4 Model evaluation against other classifiers based on accuracy
Figure 35 shows a comparison of the accuracy of our model against that of traditional models as well as other work. Our proposed LSTM model has the best performance accuracy of 0.99 when compared with traditional methods. The traditional methods, DT, K-NN and LR, showed an accuracy of 0.98, 0.96 and 0.92, respectively. Likewise, our proposed framework outperforms other techniques proposed in [11, 12, 17, 28, 29], which recorded accuracies of 0.972, 0.975, 0.987, 0.913 and 0.986, respectively.
4 Conclusion
In this paper, we developed an LSTM model for the classification of epileptic EEG signals using optimal CCP-selected features. The EEG dataset was first transformed using DWT, and 20 eigenvalues of statistical features were extracted for the experiments. CCP and PCA were applied to reduce the number of features in order to reduce the complexity of the LSTM model. Based on the results of feature reduction, 3 features were sufficient to build an effective model for epilepsy. The optimal features selected include the D1 minimum feature, D4 mean feature and A4 minimum feature. The use of these optimal features reduces the complexity of the deep LSTM model by about 13,000%. Furthermore, the model outperformed conventional models such as LR, SVM, K-NN and RNN. However, unlike PCA, CCP was found not to be suitable for GRU and BiLSTM as it resulted in lower accuracy. For future work, we suggest an investigation of the use of this CPP optimal feature selection algorithm for other neurological disorders, such as Alzheimer disease, and the development of a single model for classification of major disorders.
References
Tong Y, Aliyu I, Lim C-G (2018) Analysis of dimensionality reduction methods through epileptic EEG feature selection for machine learning in BCI. Korea Inst Electron Commun Sci 13(06):1333–1342
WHO (2018) Epilepsy. World Health Organisation
Flavio F (2016) Epilepsy. Network neuroscience. Elsevier, New York, pp 297–308. https://doi.org/10.1016/b978-0-12-801560-5.00024-0
Acharya UR, Molinari F, Sree SV, Chattopadhyay S, Ng K-H, Suri JS (2012) Automated diagnosis of epileptic EEG using entropies. Biomed Signal Process Control 7(4):401–408
Li M, Chen W, Zhang T (2017) Classification of epilepsy EEG signals using DWT-based envelope analysis and neural network ensemble. Biomed Signal Process Control 31:357–365
Bhattacharyya A, Pachori RB, Upadhyay A, Acharya UR (2017) Tunable-Q wavelet transform based multiscale entropy measure for automated classification of epileptic EEG signals. Appl Sci 7(4):385
Misiūnas AVM, Meškauskas T, Samaitienė R (2019) Algorithm for automatic EEG classification according to the epilepsy type: benign focal childhood epilepsy and structural focal epilepsy. Biomed Signal Process Control 48:118–127
Fasil O, Rajesh R (2019) Time-domain exponential energy for epileptic EEG signal classification. Neurosci Lett 694:1–8
Pedreira C, Vaudano AE, Thornton RC, Chaudhary UJ, Vulliemoz S, Laufs H, Rodionov R, Carmichael DW, Lhatoo S, Guye M (2014) Classification of EEG abnormalities in partial epilepsy with simultaneous EEG–fMRI recordings. Neuroimage 99:461–476
Sudalaimani C, Sivakumaran N, Elizabeth TT, Rominus VS (2019) Automated detection of the preseizure state in EEG signal using neural networks. Biocybern Biomed Eng 39(1):160–175
Behara DST, Kumar A, Swami P, Panigrahi BK, Gandhi TK (2016) Detection of epileptic seizure patterns in EEG through fragmented feature extraction. In: 2016 3rd international conference on computing for sustainable global development (INDIACom). IEEE, pp 2539–2542
Ahmedt-Aristizabal D, Fookes C, Nguyen K, Sridharan S (2018) Deep classification of epileptic signals. In: 2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 332–335
Hussein R, Palangi H, Wang ZJ, Ward R (2018) Robust detection of epileptic seizures using deep neural networks. In: 2018 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 2546–2550
Hussein R, Palangi H, Ward R, Wang ZJ (2018) Epileptic seizure detection: a deep learning approach. arXiv preprint arXiv:180309848
Hussein R, Palangi H, Ward RK, Wang ZJ (2019) Optimized deep neural network architecture for robust detection of epileptic seizures using EEG signals. Clin Neurophysiol 130(1):25–37
Abdelhameed AM, Daoud HG, Bayoumi M (2018) Deep convolutional bidirectional LSTM recurrent neural network for epileptic seizure detection. In: 2018 16th IEEE international new circuits and systems conference (NEWCAS). IEEE, pp 139–143
Hu X, Yuan Q (2019) Epileptic EEG identification based on deep Bi-LSTM network. In: 2019 IEEE 11th international conference on advanced infocomm technology (ICAIT). IEEE, pp 63–66
Andrzejak RG, Lehnertz K, Mormann F, Rieke C, David P, Elger CE (2001) Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys Rev E 64(6):061907
Cheah KH, Nisar H, Yap VV, Lee C-Y (2019) Convolutional neural networks for classification of music-listening EEG: comparing 1D convolutional kernels with 2D kernels and cerebral laterality of musical influence. Neural Comput Appl 3:1–25
Lee CY, Aliyu B, Lim C-G (2018) Optimal EEG locations for EEG feature extraction with application to user’s intension using a Robust neuro-fuzzy system in BCI. J Chosun Nat Sci 11(4):167–183
Güler I, Übeyli ED (2005) Adaptive neuro-fuzzy inference system for classification of EEG signals using wavelet coefficients. J Neurosci Methods 148(2):113–121
Alazrai R, Momani M, Khudair HA, Daoud MI (2017) EEG-based tonic cold pain recognition system using wavelet transform. Neural Comput Appl 5:1–14
Vishal R (2018) Feature selection-correlation and P-value. Towards Data Sci 20:20
García AMR-R, Puga JL (2018) Deciding on Null Hypotheses using P-values or Bayesian alternatives: a simulation study. Psicothema 30(1):110–115
Reddy BK, Delen D (2018) Predicting hospital readmission for lupus patients: an RNN-LSTM-based deep-learning methodology. Comput Biol Med 101:199–209
Li Z, Tian X, Shu L, Xu X, Hu B (2017) Emotion recognition from eeg using rasm and lstm. In: International conference on internet multimedia computing and service. Springer, pp 310–318
Nurujjaman M, Narayanan R, Iyengar AS (2009) Comparative study of nonlinear properties of EEG signals of normal persons and epileptic patients. Nonlinear Biomed Phys 3(1):6
Song Z, Wang J, Cai L, Deng B, Qin Y (2016) Epileptic seizure detection of electroencephalogram based on weighted-permutation entropy. In: 2016 12th world congress on intelligent control and automation (WCICA). IEEE, pp 2819–2823
Jaiswal AK, Banka H (2017) Local pattern transformation based feature extraction techniques for classification of epileptic EEG signals. Biomed Signal Process Control 34:81–92
Acknowledgements
The Basic Science Research Program supported this research through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1D1A1B03035988).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aliyu, I., Lim, C.G. Selection of optimal wavelet features for epileptic EEG signal classification with LSTM. Neural Comput & Applic 35, 1077–1097 (2023). https://doi.org/10.1007/s00521-020-05666-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-020-05666-0