Abstract
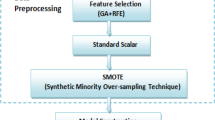
Fetal heart rate helps in diagnosing the well-being and also the distress of fetal. Cardiotocograph (CTG) monitors the fetal heart activity to estimate the fetal tachogram based on the evaluation of ultrasound pulses reflected from the fetal heart. It consists in a simultaneous recording and analysis of fetal heart rate signal, uterine contraction activity and fetal movements. Generally CTG comprises more number of features. Feature selection also called as attribute selection is a process of selecting a subset of highly relevant features which is responsible for future analysis. In general, medical datasets require more number of features to predict an activity. This paper aims at identifying the relevant and ignores the redundant features, consequently reducing the number of features to assess the fetal heart rate. The features are selected by using unsupervised particle swarm optimization (PSO)-based relative reduct (US-PSO-RR) and compared with unsupervised relative reduct and principal component analysis. The proposed method is then tested by applying various classification algorithms such as single decision tree, multilayer perceptron neural network, probabilistic neural network and random forest for maximum number of classes and clustering accuracies like root mean square error, mean absolute error, Davies–Bouldin index and Xie–Beni index for minimum number of classes. Empirical results show that the US-PSO-RR feature selection technique outperforms the existing methods by producing sensitivity of 72.72 %, specificity of 97.66 %, F-measure of 74.19 % which is remarkable, and clustering results demonstrate error rate produced by US-PSO-RR is less as well.








Similar content being viewed by others
References
Geijn HP (1996) Developments in CTG analysis. Bailliere’s Clin. Obstet. Gynaecol. 10(2):185–209
Steer PJ (2008) Has electronic fetal heart rate monitoring made a difference. Semin. Fetal Neonat. Med. 13:2–7
FIGO (1986) Guidelines for the use of fetal monitoring. Int. J. Gynecol. Obstet. 25:159–167
Gonçalves H, Rocha AP, De Campos DA, Bernardes J (2006) Linear and nonlinear fetal heart rate analysis of normal and acidemic fetuses in the minutes preceding delivery. Med. Biol. Eng. Comput. 44(10):847–855
Magenes G, Signorini MG, Arduini D (2000) Classification of cardiotocographic records by neural networks. In: Proceedings of the IEEE-INNS-ENNS international joint conference on neural networks IJCNN, vol 3, pp 637–641
Salamalekis E, Thomopoulos P, Giannaris D, Salloum I, Vasios G, Prentza A, Koutsouris D (2002) Computerised intrapartum diagnosis of fetal hypoxia based on fetal heart rate monitoring and fetal pulse oximetry recordings utilising wavelet analysis and neural networks. BJOG 109(10):1137–1142
Georgoulas G, Stylios C, Groumpos P (2005). Classification of fetal heart rate using scale dependent features and support vector machines. In: Proceedings of 16th IFAC world congress
Leski J (2003) Neuro-fuzzy system with learning tolerant to imprecision. Fuzzy Sets Syst 138:427–439
Czabanski R, Jezewski M, Wrobel J, Horoba K, Jezewski J (2008). A neurofuzzy approach to the classification of fetal cardiotocograms. In: Proceedings of 14th international conference NBC2008, vol 20, pp 446–449
Czabanski R, Jezewski M, Wrobel J, Jezewski J, Horoba K (2010) Predicting the risk of low-fetal birth weight from cardiotocographic signals using ANBLIR system with deterministic annealing and e-insensitive learning. IEEE Trans Inf Technol Biomed 14:1062–1074
Vapnik V (1998) Statistical learning theory. Wiley, New York
Vapnik V (1999) An overview of statistical learning theory. IEEE Trans Neural Netw 10:988–999
Magenes G, Signorini MG, Arduini D (2000) Classification of cardiotocographic records by neural networks. In: Proceedings of the IEEE-INNSENNS international joint conference on neural networks (IJCNN’00), vol 3, pp 637–641
Magenes G, Signorini MG, Sassi R, Arduini D (2001) Multiparametric analysis of fetal heart rate: comparison of neural and statistical classifiers. In: 9th mediterranean conference on medical and biological engineering and computing (MEDICON 2001), IFMBE Proceedings, 12–15 June 2001, Pula, Croatia, vol 1, pp 360–363
Georgoulas G, Stylios C, Bernardes J, Groumpos PP (2004) Classification of cardiotocograms using Support Vector Machines. In: Proceedings 10th IFAC symposium on large scale systems: theory and applications (LSS’04), 26–28 July 2004, Osaka, Japan
Pawlak Z (1982) Rough sets. Int J Comput Inform Sci 11(5):341–356
Nizar Banu PK, Hannah Inbarani H (2012) Performance evaluation of hybridized rough set based unsupervised approaches for gene selection. Int J Comput Intell Inf 2(2):132–141
Costa A, Ayres-de-Campos D, Costa F, Santos C, Bernardes J (2009) Prediction of neonatal acidemia by computer analysis of fetal heart rate and ST event signals. Am J Obstet Gynecol 201(5):464.e1–464.e6
Georgoulas G, Stylios CD, Groumpos PP (2006) Feature extraction and classification of fetal heart rate using wavelet analysis and support vector machines. Int J Artif Intell Tools 15(3):411–432
Georgoulas G, Stylios C, Groumpos P (2006) Predicting the risk of metabolic acidosis for newborns based on fetal heart rate signal classification using support vector machines. IEEE Trans Biomed Eng 53(5):875–884
Warrick P, Hamilton E, Precup D, Kearney R (2010) Classification of normal and hypoxic fetuses from systems modeling of intrapartum cardiotocography. IEEE Trans Biomed Eng 57(4):771–779
Chaffin DG, Goldberg CC, Reed KL (1991) The dimension of chaos in the fetal heart rate. Am J Obstet Gynecol 165(4):1425–1429
Gough NA (1993) Fractal analysis of foetal heart rate variability. Physiol Meas 14(3):309–315
Felgueiras CS, Marques de Sá JP, Bernardes J, Gama S (1998) Classification of foetal heart rate sequences based on fractal features. Med Biol Eng Comput 36(2):197–201
Kikuchi A, Unno N, Horikoshi T, Shimizu T, Kozuma S, Taketani Y (2005) Changes in fractal features of fetal heart rate during pregnancy. Early Hum Dev 81(8):655–661
Georgoulas G, Gavrilis D, Tsoulos IG, Stylios C, Bernardes J, Groumpos PP (2007) Novel approach for fetal heart rate classification introducing grammatical evolution. Biomed Signal Process Control 2(2):69–79
Van Laar J, Porath M, Peters C, Oei S (2008) Spectral analysis of fetal heart rate variability for fetal surveillance: review of the literature. Acta Obstet Gynecol Scand 87(3):300–306
Van Laar J, Peters CHL, Houterman S, Wijn PFF, Kwee A, Oei SG (2011) Normalized spectral power of fetal heart rate variability is associated with fetal scalp blood pH. Early Hum Dev 87(4):259–263
Hopkins P, Outram N, Zofgren N, Ifeachor EC, Rosen KG (2006) A comparative study of fetal heart rate variability analysis techniques. In: Proceedings of the 28th annual international conference of the ieee engineering in medicine and biology society, pp 1784–1787
Pincus SM, Viscarello RR (1992) Approximate entropy: a regularity measure for fetal heart rate analysis. Obstet Gynecol 79(2):249–255
Ferrario M, Signorini MG, Magenes G, Cerutti S (2006) Comparison of entropy based regularity estimators: application to the fetal heart rate signal for the identification of fetal distress. IEEE Trans Biomed Eng 53(1):119–125
Gonçalves H, Bernardes J, Rocha AP, Ayres-de-Campos D (2007) Linear and nonlinear analysis of heart rate patterns associated with fetal behavioral states in the antepartum period. Early Hum Dev 83(9):585–591
Ferrario M, Signorini M, Magenes G (2009) Complexity analysis of the fetal heart rate variability: early identification of severe intrauterine growth-restricted fetuses. Med Biol Eng Comput 47(9):911–919
Spilka J, Chudáček V, Koucký M, Lhotská L, Huptych M, Janku P, Georgoulas G, Stylios C (2012) Using nonlinear features for fetal heart rate classification. Biomed Signal Process Control 7(4):350–357
Questier F, Rollier IA, Walczak B, Massart DL (2002) Application of rough set theory to feature selection for unsupervised clustering. Chemometr Intell Lab Syst 63(2):155–167
Zhang J, Wang J, Li D, He H, Sun J (2003) A new heuristic reduct algorithm base on rough sets theory. LNCS, vol 2762, pp 247–253. Springer, Berlin
Swiniarski RW, Skowron A (2003) Rough set methods in feature selection and recognition. Pattern Recognit Lett 24(6):833–849
Thangavel K, Pethalakshmi A (2009) Dimensionality reduction based on rough set theory: a review. Appl Soft Comput 9(1):1–12
Echeverría JC, Άlvarez-Ramírez J, Pena MA, Rodríguez E, Gaitán MJ, González-Camarena R (2012) Fractal and nonlinear changes in the long-term baseline fluctuations of fetal heart rate. Med Eng Phys 34(4):466–471
Skinner J, Garibaldi J, Ifeachor E (1999) A fuzzy system for fetal heart rate assessment. In: Reusch B (ed) Computational intelligence. Lecture notes in computer science, vol 1625. Springer, Berlin, pp 20–29
Skinner J, Garibaldi J, Curnow J, Ifeachor E (2000) Intelligent fetal heart rate analysis. In: 1st International conference on advances in medical signal and information processing, pp 14–21
Keith R, Beckley S, Garibaldi J (1995) A multicentre comparative study of 17 experts and an intelligent computer system for managing labour using the cardiotocogram. Br J Obstet Gynaecol 102(9):688–700
Signorini M, de Angelis A, Magenes G, Sassi R, Arduini D, Cerutti S (2000). Classification of fetal pathologies through fuzzy inference systems based on a multiparametric analysis of fetal heart rate. In: Computers in cardiology, pp 435–438. Cambridge, MA
Arduini D, Giannini F, Magnes G, Signorini MG, Meloni P (2001). Fuzzy logic in the management of new prenatal variables. In: Proceedings of 5th world congress of perinatal medicine, Barcelona, vol 1, pp 1211–1216
Huang YP, Huang YH, Sandnes FE (2006) A fuzzy inference method-based fetal distress monitoring system. In: IEEE international symposium on industrial electronics, vol 1, pp 55–60, 9–13 July 2006, Montreal, Que
Hasbargen U (1994) Application of neural networks for intrapartum surveillance. In: van Geijn H, Copray F (eds) A critical appraisal of fetal surveillance. Elsevier Science (Excerpta Medica), Amsterdam, New York, pp 363–367
Beksac M, Ozdemir K, Erkmen A, Karakas U (1994) Assessment of antepartum fetal heart rate tracings using neural networks. In: van Geijn H, Copray F (eds) A critical appraisal of fetal surveillance. Elsevier Science (Excerpta Medica), Amsterdam, New York, pp 354–362
Magenes G, Signorini M G, Arduini D (2000) Classification of cardiotocographic records by neural networks. In: Proceedings IEEE-INNS-ENNS international joint conference on neural networks IJCNN, vol 3, pp 637–641
Magenes G, Signorini M, Sassi R, Arduini D (2001). Multiparametric analysis of fetal heart rate: comparison of neural and statistical classifiers. In: IFMBE proceedings of MEDICON, vol 1, pp 360–363
Noguchi Y, Matsumoto F, Maed K, Nagasawa T (2009) Neural network analysis and evaluation of the fetal heart rate. Algorithms 2:19–30
Jezewski M, Wrobel J, Horoba K, Gacek A, Henzel N, Leski J (2007) The prediction of fetal outcome by applying neural network for evaluation of CTG records. In: Kurzynski M, Puchala E, Wozniak M, Zolnierek A (eds) Computer recognition systems 2. Advances in intelligent and soft computing, vol 45. Springer, Berlin, pp 532–541
Jezewski M, Czabanski R, Wrobel J, Horoba K (2010) Analysis of extracted cardiotocographic signal features to improve automated prediction of fetal outcome. Biocybernetics and Biomedical Engineering 30:39–47
Frize M, Ibrahim D, Seker H, Walker R, Odetayo M, Petrovic D, Naguib R (2004) Predicting clinical outcomes for newborns using two artificial intelligence approaches. In: Engineering in medicine and biology society, IEMBS’04. Proceedings of 26th annual international conference of the IEEE, vol 2, pp 3202–3205
Azar AT, Nizar Banu PK, Hannah Inbarani H (2013) PSORR—An unsupervised feature selection technique for fetal heart rate. In: Proceedings of the 5th international conference on modelling, Identification and control (ICMIC 2013), Aug 31–Sept 2 2013. Cairo, Egypt, pp 60–65
Komorowski J, Pawlak Z, Polkowski L, Skowron A (1999) Rough sets: a tutorial. In: Pal SK, Skowron A (eds) Rough fuzzy hybridization: a new trend in decision making. Springer, Berlin, pp 3–98
Kalyani P, Karnan M (2011) A new implementation of attribute reduction using quick relative reduct algorithm. Int J Internet Comput 1(1):99–102
Lin TY, Cercone N (1997) Rough sets and data mining: analysis of imprecise data. Kluwer Academic Publishers, Dordrecht
Hu XT, Lin TY, Han J (2003) A new rough sets model based on database systems. In: Rough sets, fuzzy sets, data mining, and granular computing. Lecture notes in computer science, vol 2639, pp 114–121. doi:10.1007/3-540-39205-X_15
Hannah Inbarani H, Nizar Banu PK (2012) Unsupervised hybrid PSO—relative reduct approach for feature reduction. In: Proceedings of the international conference on pattern recognition, informatics and medical engineering (PRIME), pp 103–108. doi:10.1109/ICPRIME.2012.6208295
Velayutham C, Thangavel K (2011) Unsupervised feature selection using rough sets. In: Proceedings of the international conference-emerging trends in computing, pp 307–314
Velayutham C, Thangavel K (2011) Unsupervised quick reduct algorithm using rough set theory. J Electron Sci Technol 9(3):193–201
Bache K, Lichman M (2013) UCI machine learning repository. http://archive.ics.uci.edu/ml. University of California, School of Information and Computer Science, Irvine, CA
Davis JC (2002) Statistics and data analysis in geology, 3rd edn. Wiley, NewYork
Breiman L, Friedman J, Olshen R, Stone C (1984) Classification and regression trees. Wadsworth, Belmont, CA
Bridle JS (1989) Probabilistic interpretation of feedforward classification network outputs, with relationships to statistical pattern recognition. In: Fougelman-Soulie F (ed) Neurocomputing: algorithms, architectures and applications. Springer, Berlin, pp 227–236
Specht DF (1990) Probabilistic neural networks. Neural Netw 3(1):109–118
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
De Franca OF, Ferreira HM, Von Zuben FJ (2007) Applying biclustering to perform collaborative filtering. In: Proceedings of the seventh international
Xi XL, Beni G (1991) A validity measure for fuzzy clustering. IEEE Trans Pattern Anal Mach Intell 13(8):841–847
Davies DL, Bouldin DW (1979) A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 1(2):224–227
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hannah Inbarani, H., Nizar Banu, P.K. & Azar, A.T. Feature selection using swarm-based relative reduct technique for fetal heart rate. Neural Comput & Applic 25, 793–806 (2014). https://doi.org/10.1007/s00521-014-1552-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-014-1552-x