Abstract
Cardiovascular disease (CVD) is one of the leading causes of death worldwide. Early detection of CVD reduces the risk of a heart attack and increases the chance of recovery. The use of angiography to detect CVD is expensive and has negative side effects. In addition, existing CVD diagnostic methods usually achieve low detection rates and reach the best decision after many iterations with low convergence speeds. Therefore, a novel heart disease detection model based on the quantum-behaved particle swarm optimization (QPSO) algorithm and support vector machine (SVM) classification model, namely, QPSO-SVM, was proposed to analyze and predict heart disease risk. First, the data preprocessing was performed by transforming nominal data into numerical data and applying effective scaling techniques. Next, the SVM fitness equation is expressed as an optimization problem and solved using the QPSO to determine the optimal features. Finally, a self-adaptive threshold method for tuning the QPSO-SVM parameters is proposed, which permits it to drop into local minima, and balances between exploration and exploitation in the solution search space. The proposed model is applied to the Cleveland heart disease dataset and compared with state-of-the-art models. The experimental results show that the proposed QPSO-SVM model achieved the best heart-disease-prediction accuracies of 96.31% on the Cleveland heart data set. Furthermore, QPSO-SVM outperforms other state-of-the-art prediction models considered in this research in terms of sensitivity (96.13%), specificity (93.56%), precision (94.23%), and F1 score (0.95%).
Similar content being viewed by others
1 Introduction
Cardiovascular disease (CVD) is currently the leading cause of death worldwide [14]. CVDs are heart disorders caused by coronary heart disease, cerebrovascular diseases, heart failure, and other types of pathology [29]. Heart disease is mainly caused by the failure of the heart to pump enough blood into the body [1]. The most important risk factors for heart disease are an unhealthy diet, advanced age, smoking, high blood pressure, alcohol consumption, and physical inactivity. According to a World Health Organization (WHO) report [44], heart diseases kill approximately 17.9 million people each year. In the same context, the American Heart Association reported that nearly half of American adults, or approximately 121.5 million adults, are affected by CVDs [7]. Thus, it is crucial to detect cardiac factors early to effectively treat cardiac patients before a heart attack or stroke [26]. To our best knowledge, there are still few works deal with CVD prediction [13, 19]. In addition, most existing approached optimize one objective only such as model’ accuracy [27]. However, multiple conflicting objectives (such as sensitivity, specificity, precision, and F1 score) are expected to be optimized simultaneously. To that end, automated CVD prediction is one of the most important and challenging tasks globally.
Diagnostic testing and wearable monitoring are currently the two most common methods for detecting CVDs. However, extracting useful risk factors for heart disease from electronic diagnostic tests is extremely difficult because computerized medical records are unstructured and permanently expand in size [27, 32]. One obvious solution is to use a smart system, which will instantly combine the data from wearable monitoring and medical records, evaluate the data gathered, diagnose any concealed heart attack warning symptoms, and predict cardiac failure. In smart CAD diagnostic systems, Machine Learning (ML) models play a vital role due to their efficiency in classifying heart patients as normal or abnormal and predicting the output from existing data over relatively short periods to address the problem of heart disease prediction [8, 16, 36, 48]. Recenly, many ML approaches have been developed to detect the disease from training datasets containing both inputs and outputs [2, 20, 47]. For example, Naïve Bayes (NBs), Random Forest (RF) [21], Logistic Regression (LR), K-Nearest Neighbor (k-NN) [17], Support Vector Machine (SVM) [18], and Artificial Neural Networks (ANN) [10, 11] are examples of ML algorithms. These state-of-the-art models were widely commercialized and frequently enhanced by professionals in academia and industry. Although these approaches may provide the solution to the problem of heart disease detection, they are easy to drop into local extreme solutions [9]. This can be a challenge for ML models in the medical domain, where it is critical to detect heart disease with very high accuracy.
Among ML models, SVM has shown effective performance in various classification and prediction problems in many fields. Some of the current real-life applications of SVM include fault detection [45], classification of images [25], and bio-medical [5]. The use of the optimal hyperplane that separates cases of different class labels is responsible for SVM’s powerful learning capability. Moreover, SVM is sensitive to its hyperparameters, which have a direct effect on efficiency and accuracy [38].Therfore, SVM is combined with various optimization algorithms due to its strong dependence on specific parameters, such as modified binary particle swarm optimization (MBPSO) [40], genetic SVM, and analysis of variance (GSVMA) [15], gray wolf optimization (GWO) [4], genetic algorithm (GA), improved bacterial foraging optimization-based twin support vector machine (IBFO-TSVM) [30], a novel cuckoo search approach called CS-PSO-SVM [23], grasshopper optimization algorithm (GOA) [3], and ant colony optimization [34]. These hybrid ML methods have produced results that outperform conventional models.
Many optimization techniques are available in the field of swarm intelligence algorithms. Among them is the Particle Swarm Optimization (PSO) algorithm, a population-based stochastic optimization algorithm with a few parameters [12]; hence, it is simple to implement [41]. PSO is an important component in ML models because it is used to adjust SVM parameters [6]. Furthermore, PSO was used to adjust the weights of the back-propagation neural network, and it produced better results than the conventional back-propagation method [37]. Moreover, it was used to select the best feature subsets. Swarm intelligence algorithms typically identify the optimum solution when it exists, but because PSO is stochastic, they cannot be relied on to find the best solution for any given problem. Since 1995, many variants have been developed to improve the performance of the PSO and achieve good results. Liu and Fu [23] used the Chaos theory to modify the PSO inertia weight parameter. The proposed chaotic PSO balances the exploration and exploitation phases effectively. Recenly, Sun et al. [35] suggested quantum theory into PSO and developed a quantum-behaved PSO (QPSO) algorithm. QPSO achieved better results than traditional PSO, which effectively moves toward the best optimal solutions in the search space. Unlike the PSO method, the QPSO method does not require velocity vectors to move the particles and has fewer adjusting parameters overall [37]; hence, the QPSO algorithm is easier to construct than the PSO algorithm. Therefore, in this study, QPSO is uttilied to modify the optimal SVM parameter to accurately predict heart disease.
This study mainly aimed to develop a novel QPSO-SVM approach for detecting heart diseases and predicting cardiac disease. This approach takes the proprieties of SVM—simplicity, fast classification in healthcare applications, and efficiency—while avoiding its convergence to a local minimum by training SVM using a QPSO, a new version of PSO that is inspired by birds’ flock searching for the location of food. To the best of our knowledge, this study developed the first method for detecting heart illnesses using the hybrid quantum PSO and an SVM learning algorithm. The QPSO algorithm is an excellent choice for heart disease identification, and it works well on high-dimensional problems and has balanced exploitation and exploration capabilities. The first step in the QPSO-SVM model is data preparation, which includes cleaning up all datasets required to train the model and extracting information that can be used for decision-making. QPSO computes the fitness value after the parameters’ values have been adjusted. The entire swarm then evolves to generate new solutions. Subsequently, an adaptive threshold is used at the end of each generation to maintain a balance between exploitation and exploration. Finally, to evaluate the performance of the proposed QPSO-SVM, Cleveland heart disease datasets were used to build the model and compare it to some state-of-the-art methods. The QPSO-SVM outperforms other techniques in terms of classification accuracy. Furthermore, the experiments are repeated for different values of the re-adapt control parameters to show the classifiers’ sensitivity to the parameter values.
The contributions of the work are summarized as follows:
-
(1)
An improved SVM trained by a quantum-behaved PSO (QPSO) algorithm is proposed to select the optimal values of the SVM parameters and improve classification accuracy. Anovel QPSO-SVM was proposed by integrating QPSO and SVM to improve prediction accuracy.
-
(2)
The QPSO-SVM was trained and learned using public heart disease datasets to forecast the patients’ heart disease status based on their current state.
-
(3)
The proposed QPSO-SVM is evaluated and compared with the results of previous studies using evaluation metrics such as accuracy, specificity, precision, G-Mean, and F1 score. Additionally, the statistical analysis was presented to evaluate the QPSO-SVM significance compared to other models.
The remainder of this study is structured as follows: Section 2 summarizes the literature review. Section 3 describes two algorithms, SVM and QPSO. Section 4 provides a detailed explanation of the proposed QPSO-SVM model for heart disease prediction. Section 5 presents the experimental results. Section 6 contains the conclusions and recommendations for future work.
2 Literature review
This section provides a literature review of many variants of ML models used to predict heart disease. Recently, Joloudari et al. [18] suggested a new and highly efficient CAD diagnosis called GSVMA to help the effective diagnosis and prediction of CAD by selecting key features. There are two key blocks to the GSVMA approach. The first method is genetic optimization, which selects the most important features. The second is the SVM algorithm with an ANOVA kernel, which is used to categorize the input dataset. In comparison to the previously described techniques, the proposed GSVMA method performed the best on 31 of 55 features in terms of accuracy (89.45%), F-measure (80.49%), specificity (100%), sensitivity (81.22%), and area under the curve (AUC) (100%). In the same context,Perumal [28] investigated the effect of using three ML classifiers, namely, k-NN, SVM, and LR, based on the principal component analysis method. Their approach achieved 85%, 87%, and 69% accuracy rates for SVM, LR, and k-NN, respectively. It is shown that SVM and LR provide almost similar accuracy values than k-NN. Furthermore, Liu et al. [24] proposed a new model for heart disease diagnosis based on the ReliefF and Rough set (RFRS) methods. They provided two subsystems: the classification system and the RFRS feature selection system. The first system is divided into three phases: data normalization, feature extraction using the ReliefF method, and feature reduction using RFRS. The second system uses an ensemble classifier based on the C4.5 classifier. The experiments in this study were performed on the University of California Irvine (UCI) database based on the classifier C4.5. Based on a jackknife cross-validation method, it achieved a maximum classification accuracy of 92.59%. Similary, Obasi and Shafiq [8] proposed a new ML model based on existing techniques such as LR, RF, and NB classifiers. The proposed system based on the medical records of patients with 1990 observations and 18 features achieved the highest accuracy of 92.44%, 61.96%, and 59.7% for LR, RF, and NB classifiers, respectively. Besides, Latha and Jeeva [21] improved the accuracy of heart disease risk prediction based on various classifiers. An ensemble method was used, including bagging, boosting, voting, and stacking. When bagging and boosting were used, the accuracy increased by a maximum of 6.92% and 5.94%, respectively. While C4.5, PART, and Multilevel perceptrons generate an accuracy of less than 80%, the NB classifier achieves high accuracy of 83.17%.
Aljarah et al. [3] recently implemented a hybrid GOA and SVM to maximize SVM classification accuracy. The hybrid GOA-SVM was tested on 18 datasets. The experimental results are compared with GA, PSO, GWO, CS, firefly algorithm (FF), bat algorithm, and multi-verse optimizer. Although lots of efforts have been made in GOA-SVM, it has some drawbacks of being trapped in local optima. Furthermore, Vieira et al. [40] proposed MBPSO, a modified binary PSO method for detecting patients with septic shock. The experimental results show that the MBPSO achieves high accuracy compared to other PSO-based algorithms. Similary, Wei-jia et al. [43] proposed a new detection algorithm for heart diseases based on a hybrid PSO-SVM algorithm wherein PSO automatically reduces the number of features to improve SVM classifier accuracy. Experimental results are compared with other algorithms such as an ANN and feature selection-based SVM (FS-SVM). The generated rule showed that men are more likely to develop coronary heart disease than women. Subsequently, Al-Tashi et al. [4] proposed the GWO-SVM classification model for predicting heart disease. The proposed method is a combination of the feature selection method by GWO and classification by SVM. In GA-SVM, the GA is used to select the more important features, and experimental results showed that the GA-SVM model outperformed current models in terms of accuracy. When classifying heart disease using all features, the SVM achieved 83.70% accuracy. However, when classifying heart disease using the selected features, the SVM classifier achieves an accuracy of 88.34%. Liu and Fu [23] introduced a novel approach called CS-PSO-SVM for disease diagnosis depending on the hybridization of cuckoo search (CS), PSO, and SVM. CS is used as a search algorithm for finding the best initial parameter of the kernel function in SVM. Thereafter, PSO used the SVM training part to determine the best SVM parameters. The experimental results showed that CS-PSO-SVM outperforms PSO-SVM and GA-SVM in terms of classification accuracy and F-measure. However, the classification accuracy in CS-PSO-SVM still needs to be improved. Subanya and Rajalaxmi [33] developed ABC-SVM, an Artificial Bee Colony (ABC) algorithm based on swarm intelligence to identify the optimal features for heart disease detection. The SVM method is used to examine ABC’s fitness. The Cleveland heart disease dataset from the VCI ML repository is used to validate the performance of the proposed algorithm. The experimental results indicate that the ABC-SVM strategy can outperform current models in terms of classification accuracy. ABC-SVM generated an accuracy of 85.29% in the first test with five features. With seven features and the same dataset in the second experiment, it obtained an accuracy of 86.76%.
Recently, Wang et al. [42] proposed the cloud-random forest (C-RF) model, which combines the cloud model and random forest to estimate the risk of coronary heart disease. The proposed model is based on the conventional classification and regression trees (CART). It compares the C-RF model with CART, SVM, Convolutional neural network ( CNN), and RF using standard performance measures including accuracy, error rates, ROC curve, and AUC value. In comparison to CART, SVM, CNN, and RF, the C-RF model's classification accuracy is 85%, which is an improvement of 8, 9, 4, and 3%, respectively. As a result, the C-RF model performs better in terms of classification effect and performance when assessing the risk of coronary heart disease. Lin e al. [22] also used PSO to select features and determine parameter values. The SVM is then used to evaluate the classification using the selected subset. Comparing the results with those from other methods indicated that the proposed method, PSO-SVM with feature selection, performs better than PSO-SVM without feature selection. However, the experiment results of PSO-SVM must use other classifications to increase their performance. Reddy et al. [31] employed several machine learning models for efficient heart disease risk predictions such as NB, Sequential Minimal Optimization (SMO) [28], instance-based classifier (IBk), AdaBoostM1 with decision stump (DS) [24], AdaBoostM1 with LR, bagging with REPTree, bagging with LR [43], and RF. Based on the results of the experiment, SMO achieved accuracy of 85.148% using the entire set of features. Additionally, when using the chi-squared attribute evaluation method, it produced the highest accuracy of 86.468%.
Although many studies have been conducted to predict the risk of heart disease [46], they have some drawbacks such as being trapped in local optima and the need to improve the detection rate. Therefore, in this study, a novel approach, called QPSO-SVM, was proposed to effectively detect the risk of heart disease. The QPSO algorithm was used because it is simple to develop and apply, has few parameters, and produces high classification accuracy. The proposed model was applied to the Cleveland heart disease dataset that is available from the University of California, Irvine repository with two groups. Each dataset group has 303 patients and 13 attributes.
3 Preliminaries
In this section, SVM and PSO are briefly explained here.
3.1 SVM
The SVM is a well-known ML technique, developed by Vapnik in [39]. SVM maximizes the margin between the positive and negative data points closest to the decision hyperplane in an N-dimensional space when there are several classes to distinguish. The effectiveness of SVM is significantly impacted by nonlinearly separable data. However, this challenge can be overcome by transferring the data from the input space to a new, higher-dimensional space using one of the kernel functions. The objective of this kernel function is to identify the optimal decision plane.
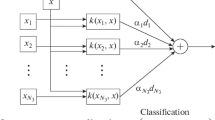
SVM is a popular ML model that is widely used in heart disease risk prediction [3,4,5, 15, 23, 25, 30, 34, 38, 40, 45]. The diagnosis of heart disease is considered an SVM classification problem that assigns the feature vector of patient \(\overrightarrow{\mathrm{x}}=\left[{\mathrm{x}}_{1},{\mathrm{x}}_{2},\dots ,{\mathrm{x}}_{\mathrm{n}}\right],\) to a class \({\mathrm{y}}_{\mathrm{j}}\in \mathrm{Y}={\{\mathrm{y}}_{1},{\mathrm{y}}_{2},\dots ,{\mathrm{y}}_{\left|\mathrm{Y}\right|}\}\) or not, where Y is a set of classes. Assume that, there are N training sets \(\left\{\left({\overrightarrow{\mathrm{x}}}_{1},{\overrightarrow{\mathrm{y}}}_{1}\right),\left({\overrightarrow{\mathrm{x}}}_{2},{\overrightarrow{\mathrm{y}}}_{2}\right),\dots ,({\overrightarrow{\mathrm{x}}}_{\mathrm{N}},{\overrightarrow{\mathrm{y}}}_{\mathrm{N}})\right\}, {\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\in {\mathrm{R}}^{\mathrm{d}}, {\mathrm{and\; y}}_{\mathrm{i}}\in (\pm 1), 1 \le \mathrm{ i }\le \mathrm{ N}\), where \({\mathrm{y}}_{\mathrm{i}},{\mathrm{y}}_{\mathrm{i}},\dots ,{\mathrm{y}}_{\mathrm{N}}\) indicate the class labels for feature vectors \(\left\{{\overrightarrow{\mathrm{x}}}_{1},{\overrightarrow{\mathrm{x}}}_{2},\dots ,{\overrightarrow{\mathrm{x}}}_{\mathrm{N}}\right\}\), respectively. In linearly separable data, the line \({\overrightarrow{\upomega }}^{\mathrm{T}}. {\overrightarrow{\mathrm{x}}}_{\mathrm{i}}+\mathrm{ b}=0\) represents the decision boundary between the two classes, positive and negative classes, where \(\overrightarrow{\upomega }\) is a weight vector, \(b\) is the bias, and \({\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\) is the input data. The goal of the SVM is to find the best parameters of \(\overrightarrow{\upomega }\) and \(\mathrm{b}\) that construct the planes \({\mathrm{H}}_{1}\mathrm{ \;and\; }{\mathrm{H}}_{2}, {\mathrm{where \;H}}_{1}\to {\overrightarrow{\upomega }}^{\mathrm{T}}. {\overrightarrow{\mathrm{x}}}_{\mathrm{i}}+\mathrm{ b}\ge +1\) for positive class and \({\mathrm{H}}_{2}\to {\overrightarrow{\upomega }}^{\mathrm{T}}. { \overrightarrow{\mathrm{x}}}_{\mathrm{i}}+\mathrm{ b}\le -1\) for the negative class, as shown in Fig. 1. Generally, SVM maximizes the margin between the positive and negative data points closest to the decision hyperplane. Here, the planes \({\mathrm{H}}_{1}\mathrm{\; and \;}{\mathrm{H}}_{2}\) can be combined as follows, \({\mathrm{y}}_{\mathrm{i}}\left({\overrightarrow{\upomega }}^{\mathrm{T}}. { \overrightarrow{\mathrm{x}}}_{\mathrm{i}}+\mathrm{ b}\right)-1\ge 0 \forall \mathrm{ i}=\mathrm{1,2},\dots ,\mathrm{N}\), where \({\mathrm{y}}_{\mathrm{i}}\in \left\{\pm 1\right\}.\) Here, the hyperplanes are formulated as an optimization problem in standard SVM, by using Eq. (1) to distinguish between the negative and positive classes, which represent the margin of SVM.
In the case of nonlinearly separable data, the standard SVM cannot be classified new cases into the correct class. The SVM introduces a kernel function (ψ) that maps the training data into a higher-dimensional space to avoid misclassification. The formulation of the objective function of SVM is given by Eq. (2):
Here, C is a penalty parameter between \({\in }_{\mathrm{i}}\) and margin size, and \({\in }_{\mathrm{i}}\) represents a slack variable.
In a nonlinear SVM classifier, the feature vector \({\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\) is labeled as \({\mathrm{i}}^{*}\) if the objective function \({\mathrm{f}}_{\mathrm{i}}\) generates the highest value for \({\mathrm{i}}^{*}\) as follows:
The results of \({\mathrm{i}}^{*}\mathrm{ th}\) objective function may be positive or negative as given in Eq. (4):
During classification, the feature vector \({\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\) that does not satisfy Eq. (4) is not classified and defined as an ambiguous case as follows:
Here, the ambiguous vectors are classified using the naïve bayes method. According to naïve bayes, the probability that the ambiguous vector \({\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\) belongs to a class \({C}_{j}\) is defined using Eq. (6):
The ambiguous vector \({\overrightarrow{\mathrm{x}}}_{\mathrm{i}}\) is labeled as \({i}^{*}\), if the conditional probability \(P({C}_{j} | { \overrightarrow{\mathrm{x}}}_{\mathrm{i}})\) is the highest for \({i}^{*}\), as given in Eq. (7):
3.2 PSO algorithm
Kennedy and Eberhart in [12] proposed the PSO optimization algorithm. A swarm in PSO is made up of a fixed number of particles. Each particle represents a single solution to the d-dimensional space optimization problem. Specifically, two types of information determine how the ith particle moves in d-dimensional space. The first type is the historically best position of the ith particle, denoted by Pbest(t) (i). The second type of information is the best among all Pbesti in the whole swarm. The global best position, denoted by gbest(t), has the highest fitness value and its velocity can be updated as follows:
where V(i) and X(i) represent the velocity and position of the ith particle in the D-th dimensional space, respectively, \(\left|\mathrm{V}(i)\right|\le {V}_{max}.\) r1 and r2 are random numbers with a uniform distribution in the range of [0, 1]. C1 and C2 are the cognition learning factors.
After evaluating the initial population, each particle in the swarm will calculate the weighted average position S(t) (i) = [S(t) (i, 1), S(t) (i, 2),... S(t) (i, d)] of their own historically best position Pbest(t) (i) and global best position gbest(t) as the attraction point and gradually move to this point. The formula for calculating the weighted average position S(t) (i, d) is as follows:
When the value of \({V}_{max}\) parameter is incorrect, particles are prevented from transitioning too quickly from breadth to depth search, causing the particle track to frequently drop into local optima. Sun, Fang, Wu, and Palade [35] proposed a quantum-behaved PSO approach to improve the performance of the PSO algorithm, which is represented by the Schrödinger equation ψ(x, t) [35], rather than position and velocity in the original PSO algorithm.
4 Proposed model
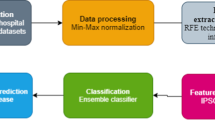
The proposed new model in this section is based on the quantum-behaved particle swarm optimization (QPSO) algorithm and SVM to analyze and diagnose heart disease risk using a real dataset. The proposed QPSO-SVM classification model consists of three phases. First, the data were processed by converting nominal data into numerical data and applying effective scaling techniques. Second, QPSO automatically adjusts the SVM parameters. Finally, the improved QPSO-SVM performs the classification tasks. Figure 2 shows the flow diagram of the proposed methodology for heart disease detection. Here, a simple and efficient hybrid model is suggesed to improve optimization capabilities without increasing the computational complexity.
4.1 Data preprocessing
Preprocessing data is the most important step before implementing the proposed model. However, real-world data cannot be directly used in the prediction task because it appears disorganized, incomplete, and contradictory. First, the categories on the training set are converted to a numerical representation, which is then applied equally to the test set. The categorical columns have values ranging from 0 to n − 1, where n is the number of categories. Next, the data is scaled from 0 to 1 using a min–max method after the missing values are replaced with random uniform noise ranging from 0 to 0.01.
4.2 Quantum-behaved particle swarm optimization algorithm
The quantum PSO (QPSO) algorithm assumes that the dynamic behavior of the particles in the PSO system meets the fundamental premise of quantum mechanics [35]. QPSO trains SVM by adjusting its parameters to avoid falling into local optima. QPSO uses the Monte Carlo method to calculate the position of the particle. The updated formula of QPSO is defined as:
where \({S}^{\left(t\right)}\left(i, d\right)\) is a random position between \(pbest\) and \(gbest\), \({u}^{\left(t\right)}(i,d)\) and \(\psi is\) a random number uniformly distributed in the range [0, 1], and \(\theta\) is an expansion coefficient.
Here, the Eqs. (10) and (12) were combined to get the generalized form of standard particle postion formula as follows:
For every ith particle, select two particles from the swarm at \(rand\left(\mathrm{0,1}\right)\) that are not the same as ith particle, respectively, k and m, and \(i\ne k\ne m,\) it is possible to calculate the difference in location between particles k and m.
Substitute Eq. (11) for \(\left({\mathrm{Pbest}}^{\left(t\right)} \left(i,d\right)-{\mathrm{ gbest}(\mathrm{d})}^{\left(t\right)}\right)\) from Eq. (10), and to increase the randomness, add \(rand\left(\mathrm{0,1}\right)\) to the second term \({\mathrm{gbest}(\mathrm{d})}^{\left(t\right)}\), and the revised evolution equation is as follows:
According to Eqs. (13) and (15), the position \({X}^{\left(t+1\right)}\left(i, d\right)\) is generated; thereafter, the individual \({X}^{\left(t+1\right)}\left(i, d\right)\) and the optimal position \({P}^{\left(t\right)}\left(i\right)\) is separated to calculate the test position T(t) (i) = [T(t) (i, 1), T(t) (i, 2),... T(t) (i, d)] as follows:
where \(\rho\) is the cross-over probability.
Using formula (13), the optimal position \({P}^{\left(t\right)}\left(i\right)\) of the ith particle is then updated:
Here, the adaptive value function is denoted by F(*). The value of \(\rho\) has a significant impact on the QPSO algorithm’s search capabilities and convergence speed. A lower \(\rho\) allows individuals in a swarm to save more information while maintaining group variety, which benefits the algorithm’s global exploration. On the other hand, higher \(\rho\) encourages individuals to learn more empirical information in the entire swarm, speeding up the algorithm’s convergence.
4.3 Hybrid QPSO-SVM
The hybrid QPSO-SVM classification model for heart disease detection is proposed. The proposed model has three stages. First, it sets the population size to N, and each ith particle position in the d-th dimensional is S(t) (i) = [S(t) (i, 1), S(t) (i, 2),... S(t) (i, d)]. Set the number of particles to M, the maximum number of iterations to \({Max}_{iter}\), and the range of particle position and particle velocity. The model uses k-fold cross-validation to ensure its effectiveness. Second, the root mean square error (RMSE) was used by the QPSO-SVM algorithm as the fitness equation. RMSE is defined as follows: RMSE = \(\sqrt{\frac{\sum_{i=1}^{N}{Y}_{i}-\widehat{{Y}_{i}}}{N}}\), where \({Y}_{i}\) and \(\widehat{{Y}_{i}}\) denote the predicated and actual values, respectively; N is the number of test samples. Thereafter, the model checks the number of iterations; if it does not equal \({Max}_{iter}\), the model updates the individual optimal position Pbest(t) (i) of the ith particle, the velocity, and the position of the ith particle using Eqs. (8) and (9), respectively. The population of the next-generation S(t + 1) (i) is as follows: S(t + 1) (i) = [S(t1) (i, 1), S(t + 1) (i, 2),... S(t + 1) (i, d)]. If \({\mathrm{Max}}_{\mathrm{iter}}\) and k-fold satisfy, it computes the average RMSE and average accuracy of k-folds. Finally, return the best optimal position Pbest, global position gbest of the whole swarm.. The fitness function value of an individual should be less than the average for a problem to be minimized, indicating that the particle's neighboring search region is potential and promising. Furthermore, the fitness function is used to evaluate the quality of individual solutions in a population of candidate solutions, and the goal of the QPSO-SVM algorithm is to evolve a population of solutions towards higher fitness values.
In any classification process, selecting a decision threshold is one of the most difficult challenges. This paper introduces a self-adaptive Threshold scheme for tuning the QPSO-SVM parameters. The SVM classification equation is formulated as an optimization problem and solved using the QPSO. The method optimizes the threshold values through effectively exploring the solution space in obtaining the global best solution. In any classification process, selecting a decision threshold is one of the most difficult challenges.
The steps of the QPSO-SVM approach are described as follows:
-
Step 1: Set the number of particles to be M, the maximum number of iteration to be \({\mathrm{Max}}_{\mathrm{iter}}\); set the range of particle position and particle velocity;
-
Step 2: The ith particle position in the d-th dimensional is S(t) (i), S(t) (i) = [S(t) (i, 1), S(t) (i, 2),..., S(t) (i, d)]; the ith particle velocity is V(t) (i), V(t) (i) = [V(t) (i, 1), V(t) (i, 2),..., V(t) (i, d)];
-
Step 3: i is set to 1;
-
Step 4: Particle position S(t) (i) is related to the QPSO-SVM algorithm for training model with a large number of samples;
-
Step 5: After that the root mean square error (RMSE) was used by QPSO-SVM algorithm uses as the fitness. RMSE is defined as follows: RMSE = \(\sqrt{\frac{\sum_{\mathrm{i}=1}^{\mathrm{N}}{\mathrm{Y}}_{\mathrm{i}}-\widehat{{\mathrm{Y}}_{\mathrm{i}}}}{\mathrm{N}}}\) (where \({\mathrm{Y}}_{\mathrm{i}}\) and \(\widehat{{\mathrm{Y}}_{\mathrm{i}}}\) denote the predicated and acual values respectively; N is the number of test samples).
-
Step 6: Update the individual optimal position Pbest(t) (i) of the ith particle;
-
Step 7: If i ≤ M, goto to step 8; otherwise, i = i + 1, go back to step 4;
-
Step 8: Update the global best value gbest(t) (i) of the ith particle;
-
Step 9: If \(\mathrm{t}\ge {\mathrm{Max}}_{\mathrm{iter}}\), go to step 12; otherwise, go to step 10;
-
Step 10: Update the velocity and position of the the ith particle using Eqs. (14) and (15) respectively. The population of the next generation S(t+1) (i) is as follows, S(t+1) (i) = [S(t1) (i, 1), S(t+1) (i, 2),..., S(t+1) (i, d)].
-
Step 11:\(\mathrm{t}=\mathrm{t}+1\), go back to step 2;
-
Step 12: Return the best optimal position pbest and global position gbest of the whole swarm.
The QPSO-SVM model is explained in detail in Algorithm 1.
4.4 Measure for performance evaluation based on self-adapting threshold
In this study, a series of operating points are generated by applying classifier thresholds to obtain a multiclass ROC curve. Here, the adaptive threshold \(\mathrm{\yen }(i,t)\) of particle i at iteration t is defined as a minimization problem as follows:
where \({\mathrm{fit}(\mathrm{gbest}(\mathrm{d})}^{\left(t\right)})\) is the best global position and fit() is the function to be optimized.
Sensitivity, specificity, accuracy, precision, G-Mean, and F-score are used in this work to assess the performance of the proposed QPSO-SVM model. Their computation requires the count of true positive and true negative at a given threshold ¥, which is calculated as follows using the confusion matrix:
-
Accuracy\(( \mathrm{\yen })\): It calculates the proportion of successfully identified cases in the test collection to the total number of observations. \({\varvec{T}}{\varvec{P}}(\mathrm{\yen })\) calculates the number of heart disease observations that the model properly classifies as heart illness at classifier threshold\(\mathrm{\yen }\). However, \({\varvec{T}}{\varvec{N}}(\mathrm{\yen })\) calculates the number of normal heart observations that the model properly classifies as the absence of heart illness at the classifier threshold\(\mathrm{\yen }\). Also, False Positive FP \((\mathrm{\yen })\) is the number of normal heart observations that the model wrongly classifies as heart disease.
$$\mathbf{A}\mathbf{c}\mathbf{c}\mathbf{u}\mathbf{r}\mathbf{a}\mathbf{c}\mathbf{y}=\frac{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })+{\varvec{T}}{\varvec{N}}(\mathrm{\yen })}{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })+{\varvec{T}}{\varvec{N}}(\mathrm{\yen })+{\varvec{F}}{\varvec{P}}(\mathrm{\yen })+{\varvec{F}}{\varvec{N}}(\mathrm{\yen })}\times 100$$(19) -
Recall\((\mathrm{\yen })\): It calculates the number of heart disease cases identified by the model divided by the total number of activities in the test set.
$$\mathbf{S}\mathbf{e}\mathbf{n}\mathbf{s}\mathbf{i}\mathbf{t}\mathbf{i}\mathbf{v}\mathbf{i}\mathbf{t}\mathbf{y}(\mathbf{R}\mathbf{e}\mathbf{c}\mathbf{a}\mathbf{l}\mathbf{l})=\frac{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })}{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })+{\varvec{F}}{\varvec{N}}(\mathrm{\yen })}\times 100$$(20) -
Specificity\((\mathrm{\yen })\): It correctly identifies people without the heart disease.
$$\mathbf{S}\mathbf{p}\mathbf{e}\mathbf{c}\mathbf{i}\mathbf{f}\mathbf{i}\mathbf{c}\mathbf{i}\mathbf{t}\mathbf{y}=\frac{{\varvec{T}}{\varvec{N}}(\mathrm{\yen })}{{\varvec{T}}{\varvec{N}}(\mathrm{\yen })+{\varvec{F}}{\varvec{P}}(\mathrm{\yen })}\times 100$$(21) -
Precision\((\mathrm{\yen })\): It calculates the amount of heart disease observations detected divided by of the total number of observations the model detect.
$$\mathbf{P}\mathbf{r}\mathbf{e}\mathbf{c}\mathbf{i}\mathbf{s}\mathbf{i}\mathbf{o}\mathbf{n}=\frac{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })}{{\varvec{T}}{\varvec{P}}(\mathrm{\yen })+{\varvec{F}}{\varvec{P}}(\mathrm{\yen })}\times 100$$(22) -
F1 Score\((\mathrm{\yen })\): It computes the weighted average of Precision rate and Recall. It is mainly used when the class distribution is unbalanced, and it is more valuable than accuracy because it calculates FP and FN.
$$\mathbf{F}1\;\mathbf{S}\mathbf{c}\mathbf{o}\mathbf{r}\mathbf{e}\left(\mathbf{\yen }\right)=2\times \frac{\mathbf{P}\mathbf{r}\mathbf{e}\mathbf{c}\mathbf{i}\mathbf{s}\mathbf{i}\mathbf{o}\mathbf{n}\left(\mathbf{\yen }\right)\times \mathbf{R}\mathbf{e}\mathbf{c}\mathbf{a}\mathbf{l}\mathbf{l}\left(\mathbf{\yen }\right)}{\mathbf{r}\mathbf{e}\mathbf{c}\mathbf{i}\mathbf{s}\mathbf{i}\mathbf{o}\mathbf{n}\left(\mathbf{\yen }\right)+\mathbf{R}\mathbf{e}\mathbf{c}\mathbf{a}\mathbf{l}\mathbf{l}\left(\mathbf{\yen }\right)}$$(23) -
G-Mean\((\mathbf{\yen })\): It is used to calculate the trade-off between sensitivity and specificity and it is an important measure for class imbalance problem.
$${\varvec{G}}-{\varvec{M}}{\varvec{e}}{\varvec{a}}{\varvec{n}}\boldsymbol{ }\left(\mathbf{\yen }\right)={\varvec{s}}{\varvec{q}}{\varvec{r}}{\varvec{t}}({\varvec{S}}{\varvec{e}}{\varvec{n}}{\varvec{s}}{\varvec{i}}{\varvec{t}}{\varvec{i}}{\varvec{v}}{\varvec{i}}{\varvec{t}}{\varvec{y}}(\mathbf{\yen })-{\varvec{S}}{\varvec{p}}{\varvec{e}}{\varvec{c}}{\varvec{i}}{\varvec{f}}{\varvec{i}}{\varvec{c}}{\varvec{i}}{\varvec{t}}{\varvec{y}}(\mathbf{\yen }))$$(24)
5 Experimental results and discussion
The proposed new classification model for heart disease detection is implemented and tested using Python 3 on a PC with an Intel (R) Core (TM) i5-7200U and RAM of 16 GB. The proposed model was ran on the heart disease dataset to investigate the feasibility of QPSO-SVM. Table 1 shows detailed information about the UCI dataset properties. The data of 297 patients shows that 137 of them have a value of 1, indicating the presence of heart disease, while the remaining 160 have a value of 0, indicating the absence of heart disease. The class value is set to 1 if the patient has cardiac issues, while 0 indicating that they do not have heart disease.
5.1 Parameters setting
Several parameters in the proposed QPSO-SVM model must be initialized before evaluation. QPSO-SVM was trained using grid search techniques. Table 2 shows the initial parameters for the competitive classification models GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVM. These parameters include the number of particles, cognition learning factors C1 and C2, mutation probabilityand other paramters.
5.2 Results of the Cleveland dataset
The effectiveness of all competitive classification algorithms, including PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVMs, is evaluated in this section using the Cleveland heart disease. The accuracy of classification models is calculated before and after threshold-offset tuning. Tables 3 and 4 show a comparison among the classification accuracies of the proposed QPSO-SVM and other classifiers from the literature for the heart disease dataset with grid search technique and using k-fold = 10. The comparison results of the classification accuracies among the proposed QPSO-SVM algorithm and the other comparative classification approaches (i.e., PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVM) are summarised in Table 3. It can be shown that QPSO-SVM achieved the highest classification accuracy under different grid parameters in comparison with other algorithms. From Table 3, it is seen that, the average accuracy achieved by ABCSVM, CS-PSO-SVM, and GOA-SVM algorithms were of 87.40%, 86.21%, and 83.98%, respectively. A slightly increased accuracy was achieved by PSO-SVM, GA–SVM, and GSVMA approaches, with an average accuracy of 88.65%, 89.28%, and 89.13%, respectively. At the same time, the GWO-SVM algorithm provides a competitive accuracy of 90.87%. However, the proposed QPSO-SVM gives an effective value with the best accuracy of 92.81%. Additionally, Table 4 displays the accuracy for the Cleveland heart disease dataset utilising classification models after threshold-offset tuning and using k-fold = 10. It shows that the average accuracy of the QPSO-SVM classifier is significant and superior to the other classifiers after threshold-offset adjustment and using k-fold = 10. QPSO-SVM has a 96.31% accuracy rate. Likewise, GA-SVM provided the second-best result with an accuracy of 93.01%. GOA-SVM, on the other hand, achieves worse outcomes and gives an accuracy rate of 87.38%. Here, to avoid local optimization, the QPSO algorithm acts in such a way that when it encounters such a location, the particles will be flied to different portions of the search space, where they will seek for optimised solutions, and this process will be repeated until the global optimised solutions are found. The technique works effectively when dealing with problems of very high dimensions as well as difficulties where the population is primarily unsuitably distributed due to the movement of the employed particles. So, the QPSO-SVM has a high success rate in addressing optimization issues.
Figure 3 illustrates the confusion matrix of correct predicted and false predicted heart detection using the proposed QPSO-SVM model and all competitive classification algorithms, including PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVMs. The proposed QPSO-SVM model has extremely few incorrect classifications, as can be seen in the figure, so the classification can be done accurately and effectively.
5.3 Comparison between different classifiers
The standard statistical p-value is used in this study to determine whether the proposed QPSO-SVM algorithm’s mean coverage values are significantly lower than those of competing models for ten k-fold scenarios. Table 5 shows that the p-values for C(QPSO-SVM, PSO-SVM), C(QPSO-SVM, GA-SVM), C(QPSO-SVM, GSVMA), C(QPSO-SVM, CS-PSO-SVM), C(QPSO-SVM, GWO-SVM), and C(QPSO-SVM, GOA-SVM) are (p < 0.00001), (p < 0.02786), (p < 0.03192), (p < 0.21138),(p < 0.000012), (p < 0.29875), and (p < 0.5507), respectively, which indicated that there is a significant difference between the performance of QPSO-SVM and PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVM for ten k-fold cases for heart disease dataset. In addaion, it is clear that the mean coverage ratios (mean) (0.400944), standard deviation (0.022743) (SD), and covariance (CV) (4.45) of C(QPSO-SVM, PSO-SVM) are superior to the ratios C(QPSO-SVM, ABC-SVM) (0.302981, 0.021531, 4.41) and C(QPSO-SVM, GSVMA) (0.270304, 0.02031, 5.45), respectively. One of the reasons why the proposed QPSO-SVM classifier outperforms previous classifiers is because it is based on our proposed adapting threshold method.
Table 6 shows the superiority of the novel QPSO-SVM algorithm. The sensitivity (96.13%), specificity (93.56%), precision (94.23%), and F1 score (0.95%) achieved by QPSO-SVM after threshold-offset tuning are better than those obtained by QPSO-SVM before threshold-offset tuning. Overall, it is seen that the QPSO-SVM method outperformed all other competing algorithms in terms of sensitivity, specificity, precision, and F1 score.
5.4 Comparison of the ROC curves of classifiers
Figure 4 shows the ROC curves and Geometric Mean or G-Mean of all classification models (i.e., PSO-SVM, GA-SVM, ABC-SVM, GWO-SVM, CS-PSO-SVM, and GOA-SVM) for the Cleveland heart disease datasets before threshold-offset tuning. The proposed QPSO-SVM model performed best (ROC = 0.91) at the start and before threshold-offset tuning on the Cleveland heart disease dataset, followed by the GA-SVM model. On the other hand, GWO-SVM had the lowest ROC score (ROC = 0.79), followed by GOA-SVM and CS-PSO-SVM models, while ABC-SVM and GSVMA performed at the average level. It can be seen that QPSO-SVM had the highest Geometric Mean or G-Mean among all classification methods (G-Mean = 90.25), followed by GSVMA (G-Mean = 89.60) and CS-PSO-SVM (G-Mean = 83.97). The reason for this result is that the QPSO-SVM model eliminates the limited particle-to-particle communication within PSO, which makes it easy to fall into a local optimum in high-dimensional space and has a slow rate of convergence during the iterative process.
Figure 5 shows the results of the ROC curve of the PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVM after threshold-offset tuning. Figure 5 also shows the effect of the preference factor of optimal threshold-offset tuning on the AUC area and G-Means. In the case of QPSO-SVM, the optimal ROC curve is around 0.37 with G-Means = 95.14 at the point of threshold-offset tuning. In contrast, PSO-SVM achieved ROC (area = 0.84) and 90.03 G-Mean at the optimal threshold of 0.21. Similarly, ABC-SVM produced 87.59 G-Mean and 0.86 ROC at a 0.35 threshold. Generally, the QPSO-SVM outperformed all other competing models in terms of G-Mean and AUC. The ABC-SVM has the worst performance in terms of G-Mean and AUC. Generally speaking, the proposed approach takes advantage of the QPSO and makes search capabilities more powerful and more stable for determining the best threshold values by utilizing the QPSO-SVM algorithm. The overall runtimes are not affected by the number of threshold values.
6 Conclusion and future work
In recent decades, great progress has been made in cardiovascular disease research. Although many studies have been conducted to address heart risk detection, most of them are ineffective and have many limitations. In this study, a hybrid model, namely, QPSO-SVM, is proposed to analyze and predict heart disease risk. The QPSO-SVM model combines the benefits of QPSO, SVM algorithms, and an adaptive threshold method. First, the data preprocessing were performed by converting nominal data into numerical data and applying effective scaling techniques. Second, the SVM parameters are automatically adjusted by QPSO. Finally, the proposed QPSO-SVM is used to classify the input data. This proposed model was evaluated by tenfold cross-validation over the Cleveland dataset, which was divided into two groups: 80% for training and 20% for testing. Furthermore, the existing state-of-the-art methods, such as PSO-SVM, GA-SVM, ABC-SVM, GSVMA, CS-PSO-SVM, GWO-SVM, and GOA-SVM, have been used to predict cardiovascular diseases based on the Cleveland dataset. Experimental results show that the proposed QPSO-SVM algorithm achieves higher accuracy of 96.31% than existing algorithms. Besides, the proposed model outperforms other heart disease prediction models considered in this research in terms of classification accuracy, specificity, precision, G-Mean, F1 score. Furthermore, the QPSO-SVM achieved good accuracy and AUC rates compared with related work.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Publicly available datasets were analyzed in this study. This data can be found here: [https://archive.ics.uci.edu/ml/datasets/heart+disease] accessed on 28 May 2023.
References
Ahmed H, Younis EM, Hendawi A, Ali AA (2020) Heart disease identification from patients’ social posts, machine learning solution on spark. Futur Gener Comput Syst 111:714–722
Akella A, Akella S (2021) Machine learning algorithms for predicting coronary artery disease: efforts toward an open source solution. Future Sci OA 7(6):FSO698. https://doi.org/10.2144/fsoa-2020-0206
Aljarah I, Al-Zoubi AM, Faris H, Hassonah MA, Mirjalili SM, Saadeh H (2017) Simultaneous feature selection and support vector machine optimization using the grasshopper optimization algorithm. Cogn Comput 10:478–495
Al-Tashi Q, Rais H, Jadid S (2019) Feature Selection Method Based on Grey Wolf Optimization for Coronary Artery Disease Classification. In: Saeed F, Gazem N, Mohammed F, Busalim A (eds) Recent Trends in Data Science and Soft Computing. IRICT 2018. Advances in Intelligent Systems and Computing, vol 843. Springer, Cham. https://doi.org/10.1007/978-3-319-99007-1_25
Babaoglu I, Findik O, Bayrak M (2010) Effects of principle component analysis on assessment of coronary artery diseases using support vector machine. Expert Syst Appl 37:2182–2185
Bashir Z, El-Hawary M (2009) Applying wavelets to short-term load forecasting using PSO-based neural networks. IEEE Trans Power Syst 24(1):20–27
Benjamin EJ et al (2018) Heart disease and stroke statistics—2018 update: a report from the American Heart Association. Circulation 137(2018):e67–e492
Chicco D, Jurman G (2020) Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Med Inform Decis Mak 20(1):16. https://doi.org/10.1186/s12911-020-1023-5
Dogan N, Tanrikulu Z (2013) A comparative analysis of classification algorithms in data mining for accuracy, speed and robustness. Inf Technol Manage 14:105–124
Dua M, Gupta R, Khari M, Crespo RG (2019) Biometric iris recognition using radial basis function neural network. Soft Comput 23(22):11801–11815
Dwivedi AK (2016) Performance evaluation of different machine learning techniques for prediction of heart disease. Neural Comput Appl 29:685–693
Eberhart R, Kennedy J (1995) A new optimizer using particle swarm theory. MHS'95. Proceedings of the Sixth International Symposium on Micro Machine and Human Science, Nagoya, Japan, p 39–43. https://doi.org/10.1109/MHS.1995.494215
García-Ordás MT, Bayón-Gutiérrez M, Benavides C, Aveleira-Mata J, Benítez-Andrades JA (2023) Heart disease risk prediction using deep learning techniques with feature augmentation. Multimed Tools Appl 82:31759–31773. https://doi.org/10.1007/s11042-023-14817-z
Ghosh P, Azam S, Jonkman M, Karim A, Shamrat FM, Ignatious E, Shultana S, Beeravolu AR, De Boer F (2021) Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 9:19304–19326
Gokulnath CB, Shantharajah SP (2018) An optimized feature selection based on genetic approach and support vector machine for heart disease. Clust Comput 22(Suppl 6):14777–14787. https://doi.org/10.1007/s10586-018-2416-4
Gupta R, Khari M, Gupta D, Crespo RG (2020) Fingerprint image enhancement and reconstruction using the orientation and phase reconstruction. Inf Sci 530:201–218
Haq AU, Li J, Memon MH, Nazir S, Sun R (2018) A hybrid intelligent system framework for the prediction of heart disease using machine learning algorithms. Mob Inf Syst 2018:3860146:1-3860146:21
Joloudari JH, Azizi F, Nematollahi MA, Alizadehsani R, Hassannataj E, Mosavi AH (2021) GSVMA: a genetic support vector machine ANOVA method for CAD diagnosis. Front Cardiovasc, 8. https://doi.org/10.3389/fcvm.2021.760178
Kishor A, Chakraborty C (2022) Artificial intelligence and internet of things based healthcare 4.0 monitoring system. Wireless Pers Commun 127:1615–1631
Kishor A, Jeberson W (2021) Diagnosis of Heart Disease Using Internet of Things and Machine Learning Algorithms. In: Singh PK, Wierzchoń ST, Tanwar S, Ganzha M, Rodrigues JJPC (eds) Proceedings of Second International Conference on Computing, Communications, and Cyber-Security. Lecture Notes in Networks and Systems, vol 203. Springer, Singapore. https://doi.org/10.1007/978-981-16-0733-2_49
Latha CB, Jeeva S (2019) Improving the accuracy of prediction of heart disease risk based on ensemble classification techniques. Inform Med Unlocked 16:100203. https://doi.org/10.1016/j.imu.2019.100203
Lin S, Ying K, Chen S, Lee Z (2008) Particle swarm optimization for parameter determination and feature selection of support vector machines. Expert Syst Appl 35:1817–1824
Liu X, Fu H (2014) PSO-based support vector machine with Cuckoo search technique for clinical disease diagnoses. Sci World J 2014:548483. https://doi.org/10.1155/2014/548483
Liu X, Wang X, Su Q, Zhang M, Zhu Y, Wang Q, Wang QA (2017) Hybrid classification system for heart disease diagnosis based on the RFRS method. Comput Math Methods Med 2017:8272091. https://doi.org/10.1155/2017/8272091
Lo C, Wang C (2012) Support vector machine for breast MR image classification. Comput Math Appl 64:1153–1162
Nandy S, Adhikari M, Balasubramanian V, Menon VG, Li X, Zakarya M (2023) An intelligent heart disease prediction system based on swarm-artificial neural network. Neural Comput Appl 35:14723–14737. https://doi.org/10.1007/s00521-021-06124-1
Obasi T, Shafiq MO (2019) Towards comparing and using Machine learning techniques for detecting and predicting Heart Attack and Diseases. IEEE Int Conf Big Data (Big Data) 2019:2393–2402
Perumal R (2020) Early prediction of coronary heart disease from cleveland dataset using machine learning techniques. Int J Adv Sci Technol 29:4225–4234
Priya L, VinilaJinny S, Mate Y (2020) Early prediction model for coronary heart disease using genetic algorithms, hyper-parameter optimization and machine learning techniques. Heal Technol 11:63–73
Rajkumar A, Bharathi A (2018) Improved bacterial foraging optimization based twin support vector machine (IBFO-TSVM) classifier for risk level classification of coronary artery heart disease in diabetic patients. Int J Appl Eng Res 13:1716–1721
Reddy K, Elamvazuthi I, Aziz AA, Paramasivam S, Chua HN, Pranavanand S (2021) Heart disease risk prediction using machine learning classifiers with attribute evaluators. Appl Sci 11(18):8352
Shanmuganathan V, Yesudhas HR, Khan MS, Khari M, Gandomi AH (2020) R-CNN and wavelet feature extraction for hand gesture recognition with EMG signals. Neural Comput Appl 32:16723–16736
Subanya B, Rajalaxmi RR (2014) Feature selection using Artificial Bee Colony for cardiovascular disease classification. Int Conf Electron Commun Syst (ICECS) 2014:1–6
Subramaniam O, Mylswamy R (2019) Ant colony optimization based support vector machine towards predicting coronary artery disease. Int J Recent Technol Eng 7:2277–3878
Sun J, Fang W, Wu X, Palade V (2012) Quantum-behaved particle swarm optimization: analysis of individual particle behavior and parameter selection. Evol Comput 20(3):349–393
Swathy M, Saruladha K (2022) A comparative study of classification and prediction of Cardio-Vascular Diseases (CVD) using Machine Learning and Deep Learning techniques. ICT Express 8:109–116
Tharwat A, Hassanien AE (2019) Quantum-behaved particle swarm optimization for parameter optimization of support vector machine. J Classif 36:576–598. https://doi.org/10.1007/s00357-018-9299-1
Tharwat A, Hassanien AE (2018) Chaotic antlion algorithm for parameter optimization of support vector machine. Appl Intell 48(3):670–686
Vapnik VN (1995) The nature of statistical learning theory. Springer, New York
Vieira SM, Mendonça LF, Farinha GJ, Sousa JM (2013) Modified binary PSO for feature selection using SVM applied to mortality prediction of septic patients. Appl Soft Comput 13:3494–3504
Wang G, Guo L (2013) A novel hybrid bat algorithm with harmony search for global numerical optimization. J Appl Math 2013:696491:1-696491:21
Wang J, Rao C, Goh MI, Xiao X (2022) Risk assessment of coronary heart disease based on cloud-random forest. Artif Intell Rev 56:203–232
Wei-jia L, Liang M, Hao C (2016) Particle swarm optimisation-support vector machine optimised by association rules for detecting factors inducing heart diseases. J Intell Syst 26:573–583
Who—cardiovascular diseases (cvds). https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 [Accessed 20 Dec 2022]
Yin S, Zhu X, Jing C (2014) Fault detection based on a robust one class support vector machine. Neurocomputing 145:263–268
Yoo H, Chung K, Han S (2020) Prediction of cardiac disease-causing pattern using multimedia extraction in health ontology. Multimed Tools Appl 80:34713–34729
Yousef R, Gupta G, Yousef N, Khari M (2022) A holistic overview of deep learning approach in medical imaging. Multimedia Syst 28:881–914
Yuvalı M, Yaman B, Tosun Ö (2022) Classification comparison of machine learning algorithms using two independent CAD datasets. Mathematics 10(3):311. https://doi.org/10.3390/math10030311
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
E.I. Elsedimy: Project administration, Resources, Validation, Conceptualization, Writing- original draft, Methodology. All experiments have done on our Faculty Labs on Egypt. Sara M. M. AboHashish: Formal analysis, Writing—review & editing. Fahad Algarni: Validation, Methodology, Software.
Corresponding author
Ethics declarations
Ethical approval
The paper does not deal with any ethical problems.
Informed consent
The authors declare that They have informed consent.
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elsedimy, E.I., AboHashish, S.M.M. & Algarni, F. New cardiovascular disease prediction approach using support vector machine and quantum-behaved particle swarm optimization. Multimed Tools Appl 83, 23901–23928 (2024). https://doi.org/10.1007/s11042-023-16194-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16194-z