Abstract
Key message
This study enriched the understanding of the mechanism of nitrogen tolerance and starvation of yellowhorn and provided a reference for the breeding of low-nitrogen tolerance germplasm in the future.
Abstract
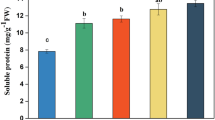
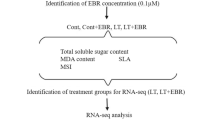
Yellowhorn is a rare woody oil crop in China, which can survive in barren, drought, cold, and even saline-alkali environments. However, its growth and development can be seriously affected by low-nitrogen stress. A comprehensive understanding of its transcriptional regulation activities under low-nitrogen stress is expected to indicate the key molecular mechanisms of its tolerance to low nitrogen levels. In this study, yellowhorn was divided into control, low-nitrogen, and nitrogen-free groups for treatment. Samples were treated for 15 days before assessing physiological characteristics and transcriptome analysis. Under low-nitrogen and no-nitrogen condition, a total of 10,733 differentially expressed genes were identified, among which 3870 genes were up-regulated and 6,863 genes were down-regulated. Under low-nitrogen stress, the most up-regulated genes were enriched in the phenylpropane synthesis pathway, flavonoid synthesis pathway, and plant hormone signal transduction pathway. Our determination of total flavonoids and proanthocyanidins also verified the upregulation of these three pathways. Brassinosteroid, salicylic, and jasmonic acid (BR, SA and JA, respectively) pathway-related genes were significantly up-regulated in the signal transduction pathway of plant hormones. This study provided a comprehensive review of the transcriptomics changes of yellowhorn under low nitrogen stress and detailed its insights into the relevant mechanism of BR, SA, and JA signaling pathway in resisting low-nitrogen stress, laying a solid foundation to further identify the corresponding molecular mechanism of yellowhorn and other woody oil plants.






Similar content being viewed by others
Data availability
The original data are stored in NCBI database, with SRA registration number: PRJNA726832 and GEO registration number: GSE223332. (https://www.ncbi.nlm.nih.gov/sra/PRJNA726832).
Abbreviations
- DEGs:
-
Differentially expressed genes
- FPKM:
-
Fragments per kb per million fragments
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- qRT-PCR:
-
Quantitative real-time PCR
- RNA-seq:
-
RNA sequence
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
- JA:
-
Jasmonates
- BR:
-
Gibberellin
- ROS:
-
Scavenging reactive oxygen species
- CK:
-
Control group
References
Babst AB, Gao F, Lucia MAG et al (2019) Three NPF genes in Arabidopsis are necessary for normal nitrogen cycling under low nitrogen stress. Plant Physiol Biochem 143:1–10. https://doi.org/10.1016/j.plaphy.2019.08.014
Ballini E, Nguyen TT, Morel JB (2013) Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 6:32. https://doi.org/10.1186/1939-8433-6-32
Begaramorales JC, Sánchezcalvo B, Chaki M et al (2015) Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J Experim Bot 66(19):5983–5996. https://doi.org/10.1093/jxb/erv306
Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206(2):522–540. https://doi.org/10.1111/nph.13269
Bi YM, Wang RL, Zhu T et al (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genom 8:281. https://doi.org/10.1186/1471-2164-8-281
Black BL, Fuchigami LH, Coleman GD (2002) Partitioning of nitrate assimilation among leaves, stems and roots of poplar. Tree Physiol 22(10):717–724. https://doi.org/10.1093/treephys/22.10.717
Bogatek R, Gniazdowska A (2007) ROS and phytohormones in plant–plant allelopathic interaction. Plant Signal Behav 2:317–318. https://doi.org/10.4161/psb.2.4.4116
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Campos ML, Yoshida Y, Major IT et al (2016) Rewiring of jasmonate and phytochrome B signaling uncouples plant growth-defense tradeoffs. Nat Commun 7:12570. https://doi.org/10.1038/ncomms12570
Chen Y, Ruberson JR, Olson DM (2008) Nitrogen fertilization rate affects feeding, larval performance, and oviposition preference of the beet armyworm, Spodoptera exigua, on cotton. Entomol Exp Appl 126:244–255. https://doi.org/10.1111/j.1570-7458.2007.00662.x
Clark ST, Verwoerd WS (2011) A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinformatics. 12(1):343. http://www.biomedcentral.com/1471-2105/12/343
Costache MA, Campeanu G, Neata G (2012) Studies concerning the extraction of chlorophyll and total carotenoids from vegetables. Romanian Biotechnol Lett 17(5):7702–7708
Cronin G, Lodge DM (2003) Effects of light and nutrient availability on the growth, allocation, carbon/nitrogen balance, phenolic chemistry, and resistance to herbivory of two freshwater macrophytes. Oecologia 137:32–41. https://doi.org/10.1007/s00442-003-1315-3
Deng B, Li YY, Lei G, Liu GH (2018) Effects of nitrogen availability on mineral nutrient balance and flavonoid accumulation in Cyclocarya paliurus. Plant Physiol Biochem 135:111–118. https://doi.org/10.1016/j.plaphy.2018.12.001
Deng B, Li YY, Lei G et al (2019) Effects of nitrogen availability on mineral nutrient balance and flavonoid accumulation in Cyclocarya paliurus. Plant Physiol Biochem 135:111–118. https://doi.org/10.1016/j.plaphy.2018.12.001
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. https://doi.org/10.1105/tpc.7.7.1085
Ebert S (2014) Potential of underutilized traditional vegetables and legume crops to contribute to food and nutritional security. Income More Sustain Production Syst Sustain 6(1):319–335. https://doi.org/10.3390/su6010319
Guo HH, Wang TT, Li QQ et al (2013) Two novel diacylglycerol acyltransferase genes from Xanthoceras sorbifolia are responsible for its seed oil content. Gene 527(1):266–274. https://doi.org/10.1016/j.gene.2013.05.076
Hsieh PH, Kan CC, Wu HY et al (2018) Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci Rep 8(1):1–23. https://doi.org/10.1038/s41598-018-30632-1
Hu S, Zhang M, Yang Y et al (2020) A novel insight into nitrogen and auxin signaling in lateral root formation in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol 20:232. https://doi.org/10.1186/s12870-020-02448-7
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18(9):477–483. https://doi.org/10.1016/j.tplants.2013.06.003
Jia J, Li S, Cao X et al (2016) Physiological and transcriptional regulation in poplar roots and leaves during acclimation to high temperature and drought. Physiol Plant 157:38–53. https://doi.org/10.1111/ppl.12400
Jin H, Zou J, Li L et al (2020) Physiological responses of yellow-horn seedlings to high temperatures under drought condition. Plant Biotechnol Rep 14:111–120. https://doi.org/10.1007/s11816-019-00590-9
Kang GZ, Wu YF, Li G et al (2019) Proteomics combined with BSMV-VIGS methods identified some N deficiency-responsive protein species and ABA role in wheat seedling. Plant Soil 444(1):177–191. https://doi.org/10.1007/s11104-019-04260-1
Karasov TL, Chae E, Herman JJ, Bergelson J (2017) Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29:666–680. https://doi.org/10.1105/tpc.16.00931
Kiba T, Kudo T, Kojima M, Sakakibara H (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Experim Bot 62:1399–1409. https://doi.org/10.1093/jxb/erq410
Kong L, Zhang Y, Du W et al (2021) Signaling responses to N starvation: focusing on wheat and filling the putative gaps with findings obtained in other plants. A review. Front Plant Sci 12:656696. https://doi.org/10.3389/fpls.2021.656696
Krapp A (2015) Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr Opin Plant Biol 25:115–122. https://doi.org/10.1016/j.pbi.2015.05.010
Lang Y, Liu Z, Zheng Z (2020) Investigation of yellow horn (Xanthoceras sorbifolia Bunge) transcriptome in response to different abiotic stresses: a comparative RNA-Seq study. RSC Adv 10:6512–6519. https://doi.org/10.1039/c9ra09535g
Li Z, Xiong F, Guo W et al (2020) The root transcriptome analyses of peanut wild species Arachis correntina (Burkart) Krapov. & W.C. Gregory and cultivated variety Xiaobaisha in response to benzoic acid and p-cumaric acid stress. Genetic Resour Crop Evol 67:9–20. https://doi.org/10.1007/s10722-019-00859-6
Lian X, Wang S, Zhang J et al (2006) Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol Biol 60:617–631. https://doi.org/10.1007/s11103-005-5441-7
Liang J, He JX (2018) Protective role of anthocyanins in plants under low nitrogen stress. Biochem Biophys Res Commun 498(4):946–953. https://doi.org/10.1016/j.bbrc.2018.03.087
Lillo C, Lea US, Ruoff P (2008) Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ 31:587–601. https://doi.org/10.1111/j.1365-3040.2007.01748.x
Lin ZH, Chen RB, Chen CS (2011) Research progress on physiological adaptability of plants to nitrogen deficiency. Hubei Agric Sci 50:4761–4764. https://doi.org/10.14088/j.cnki.issn0439-8114.2011.23.003
Liu Y, Huang Z, Ao Y et al (2013) Transcriptome analysis of yellow horn (Xanthoceras sorbifolia Bunge): a potential oil-rich seed tree for biodiesel in China. PLoS ONE 8(9):e74441. https://doi.org/10.1371/journal.pone.0074441
Liu Y, Huang Z, Ao Y et al (2014) Correction: transcriptome analysis of yellow horn (Xanthoceras sorbifolia Bunge): a potential oil-rich seed tree for biodiesel in China. PLoS ONE. https://doi.org/10.1371/annotation/803f7e8c-0718-41b4-8fc2-cc0b5f776da9
Liu DP, Li MX, Liu Y, Shi LX (2020) Integration of the metabolome and transcriptome reveals the resistance mechanism to low nitrogen in wild soybean seedling roots. Environ Experim Bot. https://doi.org/10.1016/j.envexpbot.2020.104043
Lu M, Chen M, Song J et al (2019) Anatomy and transcriptome analysis in leaves revealed how nitrogen (N) availability influence drought acclimation of Populus. Trees 33:1003–1014. https://doi.org/10.1007/s00468-019-01834-5
Luo J, Zhou J, Li H et al (2015) Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol 35(12):1283–1302. https://doi.org/10.1093/treephys/tpv091
Lv X, Zhang Y, Hu L et al (2021) Low-nitrogen stress stimulates lateral root initiation and nitrogen assimilation in wheat: roles of phytohormone signaling. J Plant Growth Regul 40:436–450. https://doi.org/10.1007/s00344-020-10112-5
Ma N, Dong L, Lü W et al (2020) Transcriptome analysis of maize seedling roots in response to nitrogen-, phosphorus-, and potassium deficiency. Plant Soil 447:637–658. https://doi.org/10.1007/s11104-019-04385-3
Meng JX, Gao Y, Han ML et al (2020) In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Hortic Plant J 6:5. https://doi.org/10.1016/j.hpj.2020.06.004
Merchante C, Alonso JM, Stepanova AN (2013) Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 16:554–560. https://doi.org/10.1016/j.pbi.2013.08.001
Nunes-Nesi A, Alisdair RF, Mark S (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3(6):973–996. https://doi.org/10.1093/mp/ssq049
Ologundudu AF, Adelusi A (2013) Effect of nitrogen nutritional stress on some growth parameters of Zea mays L. And Vigna unguiculata (L.) walp. Not Sci Biol 5(1):79–85. https://doi.org/10.15835/nsb518362
Osakabe Y, Osakabe K, Shinozaki K, Tran LS (2014) Response of plants to water stress. Front Plant Sci 5:86. https://doi.org/10.3389/fpls.2014.00086
Qi Y, Wei H, Gu W et al (2020) Transcriptome profiling provides insights into the fruit color development of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Protoplasma 258:33–34. https://doi.org/10.1007/s00709-020-01542-9
Quan X, Zeng J, Chen G et al (2019) Transcriptomic analysis reveals adaptive strategies to chronic low nitrogen in Tibetan wild barley. BMC Plant Biol 19:68. https://doi.org/10.1186/s12870-019-1668-3
Que YX, Su YC, Guo JL et al (2014) A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-Seq. PLoS ONE 9(8):e106476. https://doi.org/10.1371/journal.pone.0118445
Rajendran P, Rengarajan T, Nandakumar N et al (2014) Kaempferol, a potential cytostatic and cure for inflammatory disorders. Eur J Med Chem 86:103–112. https://doi.org/10.1016/j.ejmech.2014.08.011
Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581:2281–2289. https://doi.org/10.1016/j.febslet.2007.04.013
Ruan CJ, Yan R, Wang BX et al (2016) The importance of yellow horn (Xanthoceras sorbifolia) for restoration of arid habitats and production of bioactive seed oils. Ecol Eng 99:504–512. https://doi.org/10.1016/j.ecoleng.11.073
Sánchez FJ, Manzanares M, de Andres EF et al (1998) Turgor maintenance, osmotic adjustment and soluble sugar and proline accumulation in 49 pea cultivars in response to water stress. Field Crops Res 59:225–235. https://doi.org/10.1016/S0378-4290(98)00125-7
Sanchez-Ballesta MT, Zacarias L, Granell A, Lafuente MT (2000) Accumulation of PAL transcript and PAL activity as affected by heat-conditioning and low-temperature storage and its relation to chilling sensitivity in mandarin fruits. J Agric Food Chem 48:2726–2731. https://doi.org/10.1021/jf991141r
Sgarbi E, Fornasiero RB, Lins AP, Bonatti PM (2003) Phenol metabolism is differentially affected by ozone in two cell lines from grape (Vitis vinifera L.) leaf. Plant Sci 165:951–957. https://doi.org/10.1016/S0168-9452(03)00219-X
Sharipova G, Veselov D, Kudoyarova G et al (2016) Exogenous application of abscisic acid (ABA) increases root and cell hydraulic conductivity and abundance of some aquaporin isoforms in the ABA-deficient barley mutant Az34. Ann Bot 118:777–785. https://doi.org/10.1093/aob/mcw117
Shen Z, Zhang K, Ao Y et al (2019) Evaluation of biodiesel from Xanthoceras sorbifolia Bunge seed kernel oil from 13 areas in China. J for Res 30:869–877. https://doi.org/10.1007/s11676-018-0683-9
Solecka D, Kacperska A (2003) Phenylpropanoid deficiency affects the course of plant acclimation to cold. Physiol Plant 119:253–262. https://doi.org/10.1034/j.1399-3054.2003.00181.x
Sultana N, Islam S, Juhasz A et al (2020) Transcriptomic study for identification of major nitrogen stress responsive genes in Australian bread wheat cultivars. Front Genet 11:583785. https://doi.org/10.3389/fgene.2020.583785
Sun L, Di DW, Li G et al (2020) Transcriptome analysis of rice (Oryza sativa L.) in response to ammonium resupply reveals the involvement of phytohormone signaling and the transcription factor OsJAZ9 in reprogramming of nitrogen uptake and metabolism. J Plant Physiol 246:153137. https://doi.org/10.1016/j.jplph.2020.153137
Taiz L, Zeiger E (2002) Plant physiology, 4th edn. Sinauer Associates Inc, Sunderlan
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749. https://doi.org/10.1111/j.1365-313X.2008.03447.x
Tiwari JK, Buckseth T, Zinta R et al (2020) Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci Rep 10:1152. https://doi.org/10.1038/s41598-020-58167-4
Toma´s-Barberan FA, Gil MI, Castan˜ er M, et al (1997) Effect of Selected Browning Inhibitors on Phenolic Metabolism in Stem Tissue of Harvested Lettuce. J Agric Food Chem 45:583–589. https://doi.org/10.1021/jf960478f
Trevisan S, Begheldo M, Nonis A, Quaggiotti S (2012) The miRNA-mediated post-transcriptional regulation of maize response to nitrate. Plant Signal Behav 7:822–826. https://doi.org/10.4161/psb.20462
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86. https://doi.org/10.1186/s12870-016-0771-y
Vragovic K, Sela A, Friedlander-Shani L et al (2015) Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci U S A 112(3):923–928. https://doi.org/10.1073/pnas.1417947112
Wang Y, Chen S, Yu O (2011) Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol 91:949–956. https://doi.org/10.1007/s00253-011-3449-2
Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17(8):458–467. https://doi.org/10.1016/j.tplants.2012.04.006
Wang WS, Zhao XQ, Li M et al (2016) Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J Exp Bot 67:405–419. https://doi.org/10.1093/jxb/erv476
Wang L, Ruan CJ, Liu LG et al (2018) Comparative RNA-seq analysis of high- and low-oil yellow horn during embryonic development. Int J Mol Sci 19(10):3071. https://doi.org/10.3390/ijms19103071
Wang M, Ding YX, Wang Q et al (2020a) NaCl treatment on physio-biochemical metabolism and phenolics accumulation in barley seedlings. Food Chem 331:127282. https://doi.org/10.1016/j.foodchem.2020.127282
Wang R, Qian J, Fang Z et al (2020b) Transcriptomic and physiological analyses of rice seedlings under different nitrogen supplies provide insight into the regulation involved in axillary bud outgrowth. BMC Plant Biol 20:197. https://doi.org/10.1186/s12870-020-02409-0
Wu X, Ding C, Baerson SR et al (2018) The roles of jasmonate signalling in nitrogen uptake and allocation in rice (Oryza sativa L.). Plant Cell Environ 42(2):659–672. https://doi.org/10.1111/pce.13451. (PMID: 30251262)
Wu DG, Zhan QW, Yu HB et al (2020) Genome-wide identification and analysis of maize PAL gene family and its expression profile in response to high-temperature stress. Pak J Bot. https://doi.org/10.30848/PJB2020-5(28)
Xie YP, Chen PX, Yan Y et al (2018) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol 218(1):201–218. https://doi.org/10.1111/nph.14952
Xuan W, Beeckman T, Xu G (2017) Plant nitrogen nutrition: sensing and signaling. Curr Opin Plant Biol 39:57–65. https://doi.org/10.1016/j.pbi.2017.05.010
Yang YQ, Li X, Kong XX et al (2015) Transcriptome analysis reveals diversified adaptation of Stipa purpurea along a drought gradient on the Tibetan Plateau. Funct Integr Genom 15(3):295–307. https://doi.org/10.1007/s10142-014-0419-7
Yang Y, Wang F, Wan Q et al (2018a) Transcriptome analysis using RNA-Seq revealed the effects of nitrogen form on major secondary metabolite biosynthesis in tea (Camellia sinensis) plants. Acta Physiol Plant 40:127. https://doi.org/10.1007/s11738-018-2701-0
Yang YT, Yu Q, Yang YY et al (2018b) Identification of cold-related miRNAs in sugarcane by small RNA sequencing and functional analysis of a cold inducible ScmiR393 to cold stress. Environ Exp Bot 155:464–476. https://doi.org/10.1016/j.envexpbot.2018.07.030
Yang YY, Gao SW, Su YC et al (2019) Transcripts and low nitrogen tolerance: regulatory and metabolic pathways in sugarcane under low nitrogen stress. Environ Exp Bot 163:97–111. https://doi.org/10.1016/j.envexpbot.2019.04.010
Yao Z, Wang L, Qi J (2009) Biosorption of methylene blue from aqueous solution using a bioenergy forest waste: Xanthoceras sorbifolia seed coat. Clean 37:642–648. https://doi.org/10.1002/clen.200900093
Yao ZY, Qi JH, Yin LM (2013) Biodiesel production from Xanthoceras sorbifolia in China: opportunities and challenges. Renew Sustain Energy Rev 24:57–65. https://doi.org/10.1016/j.rser.2013.03.047
Zhang S, Zu YG, Fu YJ et al (2010) Supercritical carbon dioxide extraction of seed oil from yellow horn (Xanthoceras sorbifolia Bunge.) and its anti-oxidant activity. Bioresour Technol 101(7):2537–2544. https://doi.org/10.1016/j.biortech.2009.11.082
Zhao WC, Yang XY, Yu HJ et al (2015) RNA-seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol 56(3):455–467. https://doi.org/10.1093/pcp/pcu172
Zhao CL, Yu YQ, Chen ZJ et al (2017) Stability-increasing effects of anthocyanin glycosyl acylation. Food Chem 214:119–128. https://doi.org/10.1016/j.foodchem.2016.07.073
Zhao BB, Wang L, Pang SY et al (2020) UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Indus Crops Prod 151:112483. https://doi.org/10.1016/j.indcrop.2020.112483
Zhu L, Liu L, Sun H et al (2021) Physiological and comparative transcriptomic analysis provide nsight into cotton (Gossypium hirsutum L.) root senescence in response. Front Plant Sci 12:748715. https://doi.org/10.3389/fpls.2021.748715
Acknowledgements
We thank Sangon Biotech (Shanghai) for helping with transcriptome sequencing.
Funding
This work was financially supported by Science Technology Yuanjiang Project of Xinjiang Uygur Autonomous Region (2016E02045).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that there is no conflict of interest in this manuscript.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
468_2023_2414_MOESM2_ESM.xlsx
Supplementary file2 (XLSX 11 KB) Table S1. Sample QC data information statistics. Data are presented as the means ± standard error. Asterisk indicates statistically significant difference (p < 0.05).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, X., Zou, J., Jin, C. et al. Transcriptomics analysis reveals Xanthoceras sorbifolia Bunge leaves’ adaptation strategy to low nitrogen. Trees 37, 1153–1166 (2023). https://doi.org/10.1007/s00468-023-02414-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-023-02414-4