Abstract
Key message
The selected material of Cerasus subgen. will be useful for conservation and management and important for Prunus breeding programs.
Abstract
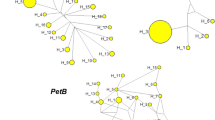
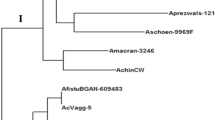
Knowledge of relationships among the cultivated and wild species of Cerasus is important for recognizing gene pools in germplasm and developing effective conservation and management strategies. In this study, genetic and phylogenetic relationships of wild Cerasus subgenus species naturally growing in Iran, including P. avium (mazzard), P. mahaleb, P. brachypetala, P. incana, P. yazdiana, P. microcarpa subsp. microcarpa, P. microcarpa subsp. diffusa and P. pseudoprostrata and three commercial species, sweet cherry (P. avium), sour cherry (P. cerasus) and duke cherry (P. x gondouinii) was investigated based on 16 nuclear SSR and five chloroplast SSR. Very high level of polymorphism was detected among the studied species based these molecular markers, indicating high inter and intraspecific genetic variation. Inter and intraspecific genetic similarity coefficients varied from 0.00 to 1.00, indicating high genetic variation in studied germplasm. These two molecular markers types could distinguish differences between all species so that accessions of each species were placed into a single group. Based on molecular markers, a close correlation was observed between intraspecific variation and geographical distribution. Furthermore, based on nuSSR primers, most wild species showed 2–4 alleles and may be tetraploid. In conclusion, the conservation of these highly diverse native populations of Iranian wild Cerasus germplasm is recommended for future breeding activity.





Similar content being viewed by others
References
Antonius K, Aaltonen M, Uosukainen M, Hurme T (2012) Genotypic and phenotypic diversity in Finnish cultivated sour cherry (Prunus cerasus L.). Genet Resour Crop Evol 59:375–388
Aranzana MJ, Garcia-Mas J, Carbo J, Arus P (2002) Development and variability analysis of microsatellite markers in peach. Plant Breed 121:87–92
Arnold ML, Robinson JJ, Buckner CM, Bennet BD (1992) Pollen dispersal and interspecific gene flow in Louisiana irises. Heredity 68:399–404
Avramidou I, Ganopoulos V, Aaavanopoulos FA (2010) DNA fingerprinting of elite Greek wild cherry (Prunus avium L.) genotypes using microsatellite markers. Forestry 83:527–533
Beaver JA, Iezzoni AF, Ramm CW (1995) Isozyme diversity in sour, sweet and ground cherry. Theor Appl Genet 90:847–852
Birky CW (1988) Evolution and variation in plant chloroplast and mitochondrial genomes. In: Gottlieb L, Jain S (eds) Plant evolutionary biology. Chapman and Hall, London, pp 23–53
Bouhadida M, Martin JP, Eremin G, Pinochet J, Moreno MA, Gorgocena Y (2007) Chloroplast DNA diversity in Prunus and its implication on genetic relationships. J Am Soc Hortic Sci 132:670–679
Bouhadida M, Casas AM, Gonzalo MJ, Arus P (2009) Molecular characterization and genetic diversity of Prunus rootstocks. Sci Hortic 120:237–245
Brettin TS, Karle R, Crowe EL, Iezzoni F (2000) Chloroplast inheritance and DNA variation in sweet, sour and ground cherry. Heredity 91:74–79
Cantini C, Iezzoni A, Lamboy AF, Boritzki M, Struss D (2001) DNA fingerprinting of tetraploid cherry germplasm using simple sequence repeats. J Am Soc Hortic Sci 126:205–209
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Clarke JB, Tobutt KR (2003) Development and characterization of polymorphic microsatellites from Prunus avium ‘Napoleon’. Mol Ecol Notes 3:578–580
Clarke JB, Tobutt KR (2009) A Standard Set of Accessions, Microsatellites and Genotypes for Harmonising the Fingerprinting of Cherry Collections for the ECPGR. Acta Hortic 814:615–619
Decroocq V, Hagen LS, Favé MG, Eyquard JP, Pierronnet A (2004) Microsatellite markers in the hexaploid Prunus domestica species and parentage lineage of three European plum cultivars using nuclear and chloroplast simple-sequence repeats. Mol Breed 13:135–142
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arus P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Downey SL, Iezzoni AF (2000) Polymorphic DNA markers in black cherry (Prunus serotina) identified using sequences from sweet cherry, peach, and sour cherry. J Am Soc Hortic Sci 125:76–80
Ercisli S, Agar G, Yildirim N, Duralija B, Vokurka A, Karlidag H (2011) Genetic diversity in wild sweet cherries (Prunus avium) in Turkey revealed by SSR markers. Genet Mol Res 10:1211–1219
Freimer NB, Slatkin M (1996) Microsatellites: evolution and mutational processes. Variation in the human genome. Ciba Foundation Symposium, vol 197. Ciba Foundation, London, pp 51–72
Ganopoulos IV, Avramidou E, Fasoula DA, Diamantidis G, Aravanopoulos FA (2010) Assessing inter- and intra-cultivar variation in Greek Prunus avium by SSR markers. Plant Genet Res: Charact Util 8:242–248
Haider A (2011) Chloroplast-specific universal primers and their uses in plant studies. Biol Plant 55:225–236
Harris SA, Ingram R (1991) Chloroplast DNA and biosystematics—the effects of intraspecific diversity and plastid transmission. Taxon 40:393–412
Heinze B (1999) Molecular genetic investigations in wild and cultivated Prunus avium in Austria and beyond. In: Ritter E, Espinel S (eds) Proceedings of applications of biotechnology to forest genetics. International Congress, Vitoria-Gasteiz, pp 77–80
Hormaza JI (2002) Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor Appl Genet 104:321–328
Horvath A, Zanetto A (2008) Origin of sour cherry (Prunus cerasus L.) genomes. Acta Hortic 795:131–136
Iezzoni AF, Schmidt H, Albertini A (1990) Cherries (Prunus). In: Moore JN, Ballington JR Jr (eds) Genetic resources of temperate fruit and nut crops, vol 1. ISHS, Wageningen, pp 111–173
Khadivi-Khub A, Zamani Z, Fatahi MR (2012) Multivariate analysis of Prunus subgen. Cerasus germplasm in Iran using morphological variables. Genet Resour Crop Evol 59:909–926
Ma H, Olsen R, Pooler M (2009) Evaluation of flowering cherry species, hybrids, and cultivars using simple sequence repeat markers. J Am Soc Hortic Sci 134:435–444
Mantel NA (1967) The detection of disease clustering gene and a generalized regression approach. Cancer Res 27:209–220
Mariette S, Tavaud M, Arunyawat U, Capdeville G, Millan M, Salin F (2010) Population structure and genetic bottleneck in sweet cherry estimated with SSRs and the gametophytic self-incompatibility locus. BMC Genet 11:77–86
Matus IA, Hayes PM (2002) Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. Genome 45:1095–1106
Mohanty A, Martin JP, Aguinagaldo I (2001a) Chloroplast DNA study in wild population and some cultivars of Prunus avium. Theor Appl Genet 103:112–117
Mohanty A, Martinn JP, Aguinagalde I (2001b) A population genetic analysis of chloroplast DNA in wild populations of Prunus avium L. in Europe. Heredity 87:421–427
Mohanty A, Martin JP, Gonzaalez LM, Aguinagaldo I (2003) Association between chloroplast DNA and mitochondrial DNA haplotypes in Prunus spinosa L. (Rosaceae) populations across Europe. Ann Bot 92:749–755
Mozaffarian V (2002) Studies on the flora of Iran, new species and new records. Pak J Bot 34:391–396
Nas MN, Bolek Y, Adem B (2011) Genetic diversity and phylogenetic relationships of Prunus microcarpa C.A. Mey. subsp. tortusa analyzed by simple sequence repeats (SSRs). Sci Hortic 127:220–227
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Ohta S, Nishitani C, Yamamoto T (2005a) Chloroplast microsatellites in Prunus, Rosaseae. Mol Ecol Notes 5:837–840
Ohta S, Katsuki T, Tanaka T, Hayashi T, Sato Y, Yamamoto T (2005b) Genetic variation in flowering cherries (Prunus subgenus Cerasus) characterized by SSR markers. Breed Sci 55:415–424
Ohta S, Yamamoto T, Nishitani C, Katsuki T, Iketani H (2007) Phylogenetic relationships among Japanese flowering cherries (Prunus subgenus Cerasus) based on nucleotide sequences of chloroplast DNA. Plant Syst Evol 263:209–225
Olden EJ, Nybom N (1968) On the origin of Prunus cerasus L. Heredity 59:327–345
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Palmer JD (1987) Chloroplast DNA evolution and biosystematics uses of chloroplast DNA variation. Am Nat 130:S6–S29
Panda S, Martin JM, Aguinagalde I (2003a) Chloroplast DNA study in sweet cherry cultivars using PCR-RFLP method. Gen Res Crop Evol 50:489–495
Panda S, Martin JP, Aguinagalde I, Mohanty A (2003b) Chloroplast DNA variation in cultivated and wild Prunus avium L.: a comparative study. Plant Breed 122:92–94
Pavlicek A, Hrda S, Flegr J (1999) FreeTree—Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Folia Biol (Praha) 45:97–99
Powell W, Morgante M, McDevitt R, Vendramin GC, Rafalski JA (1996) Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA 92:7759–7763
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147
Rechinger KH (1969) Rosaceae. In: Rechinger KH (ed) Flora Iranica, vol 66. Akademische Druck-U, Austria, pp 187–203
Rehder A (1940) Manual of cultivated trees and shrubs hardy in North America exclusive for subtropical and warmer temperate regions, 2nd edn. Macmillan, New York
Rohlf FJ (2000) NTSYS-pc Numerical Taxonomy and Multivariate Analysis System. Version 2.1. Exeter Software, Setauket, NY
Romesburg HC (1990) Cluster Analysis for Researchers. Krieger Publishing Company, Malabar
Santi F, Lemoine M (1990) Genetic markers for Prunus avium L. 2. Clonal identifications and discrimination from P. cerasus and P. cerasus × P. avium. Annales des Sciences Forestières 47:219–227
Schueler S, Tusch A, Schuster M, Ziegenhagen B (2003) Characterization of microsatellites in wild and sweet cherry (Prunus avium L.) markers for individual identification and reproductive processes. Genome 46:95–102
Sefc KM, Lopez MS, Lefort F, Botta R (2000) Microsatellites variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet 100:498–505
Sneller CH, Miles JW, Hoyt JM (1997) Agronomic performance of soybean plant introductions and their genetic similarity to elite lines. Crop Sci 37:1595–1600
Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG (2000) Characterisation of microsatellite markers in peach [Prunus persica (l.) Batsch]. Theor Appl Genet 101:421–428
Steinkellner H, Lexer C, Turetschek E, Glossl J (1997) Conservation of (GA)n microsatellite loci between Quercus species. Mol Ecol 6:1189–1194
Struss D, Boritzki M, Karle R, Iezzoni AF (2002) Microsatellite markers differentiate eight Giessen cherry rootstocks. Hortic Sci 37:191–193
Struss D, Ahmad R, Southwick SM, Boritzki M (2003) Analysis of sweet cherry (Prunus avium L) cultivars using SSR and AFLP. J Am Soc Hortic Sci 128:904–909
Tavaud M, Zanetto A, David JL, Laigret F, Dirlewanger E (2004) Genetic relationships between diploid and allotetraploid cherry species (Prunus avium, Prunus x gondouinii and Prunus cerasus. Heredity 93:631–638
Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, Pancaldi M, Sansavini S (2000) Microsatellite DNA in peach (Prunus persica (L.) Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43:512–520
Turkec A, Muge S, Berthold H (2006) Identification of sweet cherry cultivars (Prunus avium L.) and analysis of their genetic relationship by chloroplast sequence-characterized amplified regions (cpSCAR). Genet Res Crop Evol 53:1653–1654
Turkoglu Z, Bilgener S, Ercisli S, Bakir M, Koc A, Akbulut M, Gercekcioglu R, Gunes M, Esitken A (2010) Simple sequence repeat-based assessment of genetic relationships among Prunus rootstocks. Genetics Mol Res 9:2156–2165
Tzedakis PC, Lawson IT, Frogley MR, Hewitt GM (2002) Buffered tree population changes in Quaternary refugium: evolutionary implication. Science 292:267–269
Vaughan SP, Russell K (2004) Characterization of novel microsatellites and development of multiplex PCR for large-scale population studies in wild cherry, Prunus avium. Mol Ecol Notes 4:429–431
Watkins R (1976) Cherry, plum, peach, apricot and almond. In: Simmonds NW (ed) Evolution of crop plants. Longman, London, UK, pp 242–247
Wen J, Berggren ST, Lee CH, Ickert S (2008) Phylogenetic inferences in Prunus (Rosaceae) using chloroplast ndhF and nuclear ribosomal ITS sequences. J Syst Evol 46:322–332
Wünsch A (2009) Cross-transferable polymorphic SSR loci in Prunus species. Sci Hortic 120:348–352
Wünsch A, Hormaza JI (2002) Molecular characterisation of sweet cherry (Prunus avium L.) genotypes using peach (Prunus persica (L.) Batsch) SSR sequences. Heredity 89:56–63
Wünsch A, Hormaza JI (2004) Molecular evaluation of genetic diversity and S-allele composition of local Spanish sweet cherry (Prunus avium L.) cultivars. Genet Resour Crop Evol 51:635–641
Xuan H, Neumueller M, Schlottmann P (2010) Approaches to determine the origin of European plum (Prunus domestica) based on nucleotide sequences of chloroplast DNA. In: Proceedings of the 28th international horticultural congress, Lisboa, Portugal, 22–27 August
Zeinalabedini M, Majourhat K, Khayam-Nekoui M, Grigorian V, Torchi M, Dicenta F, Martinez-Gomez P (2008) Comparison of the use of morphological, protein and DNA markers in the genetic characterization of Iranian wild Prunus species. Sci Hortic 116:80–88
Zeinalabedini M, Khayam-Nekoui M, Grigorian V, Gradziel TM, Martinez-Gomez P (2010) The origin and dissemination of the cultivated almond as determined by nuclear and chloroplast SSR marker analysis. Sci Hortic 125:593–601
Acknowledgment
We thank the Ministry of Science, Research and Technology of the Islamic Republic of Iran and Center of Excellence for Temperate Fruit Research in University of Tehran for financial assistance. We also thank CITA (Centro de Investigación y Tecnología Agroalimentaria de Aragón), Zaragoza, in particular, Fruit-Tree Department, for providing the facilities to do this research. We gratefully acknowledge Teresa Bespín for excellent assistance of technical markers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Canovas.
Rights and permissions
About this article
Cite this article
Khadivi-Khub, A., Zamani, Z., Fattahi, R. et al. Genetic variation in wild Prunus L. subgen. Cerasus germplasm from Iran characterized by nuclear and chloroplast SSR markers. Trees 28, 471–485 (2014). https://doi.org/10.1007/s00468-013-0964-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-013-0964-z