Abstract
Feeding [1-14C]-guanidinoacetic acid to shoot primordia, O2 uptake was inhibited and major products were 14C-glycine, 14CO2 and 14C-serine. The direct decarboxylation of [1-14C]-guanidinoacetic acid to 14CO2 and N-methylguanidine, the methylation of [1-14C]-guanidinoacetic acid to 14C-creatine, and the lytic cleavage to urea and 14C-glycine were all ruled out. Enzymatic transamidinations of [1-14C]-guanidinoacetic acid with amino acid acceptors occurred as arginine-rich storage proteins were being turned over and new proteins synthesized containing 14C-glycine and 14C-serine. The products of transamidination were recycled as substrates until 14C-glycine was metabolized in different directions and transported to mitochondria and peroxisomes. 14C-Glycine was decarboxylated by a glycine decarboxylase multienzyme complex resulting in a net carbon loss and a sharp decline in total protein rich in arginine N. Under these conditions, unlabelled arginine and ornithine contributed as substrates for reversible transamidination reactions. Peroxisomes and mitochondria are hypothesized as providing arginine-derived nitric oxide to maintain redox homeostasis in response to the stresses imposed by [1-14C]-guanidinoacetic acid and to protect against the inhibitory activity of sulfhydryls on transamidinase activity. The destruction of a respiratory inhibitor by transamidination may comprise a mechanism associated with the awakening from of dormancy and the mobilization of storage protein reserves in conifers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Following the isolation of arginine from conifers by Schulze (1896) its physiological role and fate were difficult to explain (Mothes 1929). The formation of arginine-derived guanidino compounds in white spruce [Picea glauca (Moench) Voss] was demonstrated with [UL-14C]-l-arginine, [amidine-14C]-l-arginine, and [1,2,3,4-14C]-γ-guanidinobutryic acid (Durzan 1968, 1969a). The transamidination of 1[UL-14C]-l-arginine was postulated as explaining some of the several guanidino compounds derived from arginine, one of which co-chromatographed with guanidinoacetic acid (glycocyamine). Transamidination transfers an amidine group (NH2C=NH) from one guanidino compound to an acceptor having an amino group (Meister 1965; van Thoai 1965).
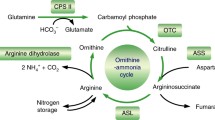
The availability of [1-14C]-guanidinoacetic acid enabled four hypotheses in Fig. 1 to be tested under aseptic conditions as excised vegetative shoot primordia entered winter dormancy (Fig. 2). First, the decarboxylation of [1-14C]-guanidinoacetic acid produces N-methylguanidine and 14CO2. Second, the methylation of [1-14C]-guanidinoacetic acid yields 14C-creatine, creatinine and possibly N-methylglycine (sarcosine). Creatine has been reported in wheat (Barrenscheen and Pany 1941; Barrenscheen and von Vály-Nagy 1942), but its natural occurrence in plants has not been verified. In humans, guanidinoacetic acid is methylated to creatine (Wyse and Kaddurah-Daouk 2000). Third, a guanidinoacetase (glycocyaminase) yields 14C-glycine and urea from [1-14C]-guanidinoacetic acid and water (Brunel et al. 1967; Roche et al. 1950; Yorifuji et al. 1977). This enzyme occurs in microorganisms and is not known in plants.
The fate of carbon (black dot) in [1-14C]-guanidinoacetic acid was based on four postulated enzymatic reactions occurring within 32 h in shoot primordia entering winter dormancy. One is the decarboxylation of [1-14C]-guanidinoacetic acid to N-methylguanidine and 14CO2. Another is the formation of urea and 14C-glycine by contaminating microorganisms having guanidinoacetases. The third deals with the biosynthesis of 14C-creatine from [1-14C]-guanidinoacetic acid requiring methylation of guanidinoacetic acid via S-adenosylmethionine (AdoMet) with the formation of S-adenosylhomocysteine (AdoHcy). No evidence was found for all three reactions under aseptic conditions. The fourth hypothesis requires the transfer (transamidination) the amidine group (NH2C=NH) from [1-14C]-guanidinoacetic acid to an acceptor having an amino group. Transamidination of [1-14C]-guanidinoacetic acid with ornithine (orn), derived from the turnover of arginine-rich proteins, yielded 14C-glycine and regenerated free arginine (arg). Recycling of [1-14C]-guanidinoacetic acid and 14C-glycine by transamidination continued as 14C-glycine was decarboxylated to 14CO2 by the glycine decarboxylase mitochondrial complex (GDMC) or metabolized to 14C-serine and traces of 14C-glutamic acid
Proteins stained by fast-green in a longitudinal section of a dormant white spruce bud. M is the new meristematic vegetative shoot, m is a cavity filled with liquid below the crown CR also called a nodal diaphragm or collenchyma plate. S bud scales, lp leaf primordia, r resin duct. Vertical arrow at left is 2 mm. The upper horizontal arrow indicates the plane of excision of the shoot primordium for exposure to guanidinoacetic acid under aseptic conditions. The lower arrow indicates the cut made to collect the buds under field conditions
Fourth, the reversibility and wide range of substrates for transamidinase activity offer several possibilities (Meister 1965; van Thoai 1965). The transamidination of [1-14C]-guanidinoacetic acid with ornithine as an acceptor of the amidino group will yield arginine and 14C-glycine. When [1-14C]-guanidinoacetic acid transfers its amidino moiety to glycine, the products are guanidinoacetic acid and 14C-glycine. If arginine is an amidino donor, and glycine or 14C-glycine are the amidino acceptors, the products are guanidinoacetic or [1-14C]-guanidinoacetic acid, and ornithine, respectively. This study investigates which of the four hypotheses would explain the fate of [1-14C]-guanidinoacetic acid in shoot primordia of buds entering winter dormancy.
Materials and methods
White spruce buds were collected on November 6 from a 27-year old tree exposed to full natural light at the Petawawa Forest Experiment Station (45o08′N, 81o27′W). Shoot primordia, being enclosed by bud scales and entering winter dormancy, were not photosynthetically active. Vegetative shoot primordia with their crowns were easily separated from bud scales and shoot branches. Proteins (Fig. 2) were stained at pH 2.2 with fast-green (Woodard et al. 1961). In spruce buds, their “crowns” isolate shoot primordia from the mature underlying tissue except where penetrated by vascular traces (Romberger 1963). The parenchyma below the crown breaks down in autumn to form a cavity partially filled with viscous material. It was not used in this study.
Shoot collections were made in the field between 8 and 8.30 a.m. at temperatures between 15 and 17°C to minimize diurnal variations in amino acid content. In the laboratory, buds on shoots were kept at 20 ± 3°C for the removal of bud scales and to isolate shoot primordia within 1 h of collection (Fig. 2). Shoot primordia were surface sterilized in 5% (v/v) Chlorox, and rinsed in sterile water for duplicated experiments in Warburg flasks at 20 ± 1°C.
To each pre-autoclaved flask of a Warburg apparatus, 25 shoot primordia of uniform size (each 9 ± 2 mg f wt) were added with their crowns facing downward to 2.5 ml of sterile-filtered 0.01 M phosphate buffer at pH 6.8. With separate batches of 25 shoot primordia, the relative turgidity (RT), based on 10 mg dry wt, was measured at 1, 4, 8, 16, and 32 h (Weatherley and Slatyer 1957).
Sterile-filtered [1-14C]-guanidinoacetic acid was added at 185 × 103 Bq (sp. act. 618 × 105 mM) to each flask containing 10% KOH in filter paper in the center well. From the time of shoot excision to the end of the study at 32 h, cool white incandescent light did not exceed 0.2 mmol/(m2 s). Flasks were shaken at 10 rpm at a radius of 2.5 cm. Oxygen uptake was measured at 30-min intervals in flasks with and without [1-14C]-guanidinoacetic acid for the full 32 h of the experiment. All shoot primordia from each duplicated treatment and controls were collected at 4, 8, 16, and 32 h for biochemical analyses.
Shoot primordia were immediately fixed and extracted with 70% (v/v) ethanol, and homogenized for the extraction of the soluble N. After five extractions, a sixth extraction was tested with ninhydrin and by scintillation counting to verify that all the amino acid N and isotopically labeled products were extracted. The homogenization of primordia with [1-14C]-guanidinoacetic acid and 70% ethanol at zero time yielded no metabolic products. After the experiment, five primordia from controls and treatments with [1-14C]-guanidinoacetic acid were transferred to plates containing nutrient-rich agar to check for contamination. None showed any contamination over a subsequent 3-week period.
Two independent analytical systems were used to profile the metabolic fate of [1-14C]-guanidinoacetic acid. One was two-dimensional paper chromatography with autoradiography (Durzan 1968). The other was automated amino acid analysis for physiological fluids using ion-exchange chromatography with a Beckman Amino Acid Analyzer, model 120C (Benson and Patterson 1965). The latter was modified for flow-cell scintillation counting for carbon-14, fraction collection, and for the quantitative analyses of guanidino compounds using the Sakaguchi reagent (Durzan 1969b). Both systems independently confirmed the identities of the metabolites derived from [1-14C]-guanidinoacetic acid.
Ethanolic extracts (70% v/v) were passed through Dowex 50 × 4 (200 mesh) columns to remove the neutral and anionic fractions and improve the separation of amino acids on paper chromatograms and autoradiographs. The cation fractions, containing the free-amino acids and guanidino compounds, were eluted with 4 M NH4OH for analyses by paper chromatography and on the amino acid analyzer. All column regeneration fractions were kept for metabolic profiling.
For paper chromatograms, the identities and locations of the amino acids agreed with pure standards and by their reactions with ninhydrin, UV light and non-reaction with the Sakaguchi reagent. The differential color reaction of [1-14C]-guanidinoacetic acid with the Sakaguchi reagent agreed with an authentic guanidinoacetic acid not containing carbon-14. The identification of amino acids and guanidino compounds containing radioactivity was based on the commercially available internal standards for physiological fluid analyses (Benson and Patterson 1965; Durzan 1969b). The alcohol-insoluble residue was kept for acid hydrolysis to recover the radioactivity and determine the N content in protein amino acids. Free and protein amino acids were quantitatively determined on the amino acid analyzer within ±3% as previously described (Durzan 1968).
The NaOH fractions, used to strip and regenerate the ion-exchange columns of the amino acid analyzer, were collected and monitored for 14C-guanidine and 14C-N-methylguanidine using commercially available 14C-guanidine and non-radioactive N-methylguanidine as controls. The 14CO2 trapped in KOH and in 14C-metabolites on paper chromatograms or from the amino acid analyzer were determined with 70–80% efficiency by a liquid scintillation counter using a channels ratio method for quench correction to 100% (Becker 1967). In Table 1, the means of duplicated values are within for all 12% and all values are based on 100 mg f wt.
Results
At 4 h, the RT of excised shoot primordia in phosphate buffer was 25 ± 3% when the uptake of [1-14C]-guanidinoacetic acid reached 1,722 Bq/100 mg f wt. At 16 h, the RT increased to 27 ± 4%. By 32 h, the RT was 36 ± 6 %. When compared with the 4 h exposure, the levels of [1-14C]-guanidinoacetic acid in the soluble N pool at 32 h remained steady and increased only by 58 Bq (Table 1). Steady levels of [1-14C]-guanidinoacetic acid would be maintained by transamidinase activities, which recycle 14C-glycine and [1-14C]-guanidinoacetic acid as substrates.
Over time, the ratio of 14CO2 released to O2 consumed was considerably lower than 1. This indicated that protein and fats, which give a respiratory quotient less than 1, were being oxidized. Between 4 and 32 h, the total protein N declined from 742 to 268 μg/100 mg f wt. The 14CO2 trapped by KOH increased nearly fourfold compared with the [1-14C]-guanidinoacetic acid in the free-amino acid pool. Most 14CO2 originated from 14C-glycine and 14C-serine rather than from the organic acid fraction (Table 1; Fig. 3).
The amino acid pool (excluding [1-14C]-guanidinoacetic acid), contained mainly 14C-glycine and 14C-serine. Low levels of 14C-glutamic acid and 14C-proline were detected on the amino acid analyzer and not picked up on autoradiographs unless longer exposure times were used. The low levels of 14C-organic acids indicated that most of the release 14CO2 originated from the decarboxylation of 14C-glycine.
The specific activity of 14C-glycine in the free-amino acid pool declined significantly over time. This coincided with the steady drop in microgram of total protein N (Table 1). Protein turnover provided unlabeled glycine lowering the specific activities of the carbon in 14C-glycine. Newly synthesized 14C-glycine and unlabeled glycine were used for protein synthesis even as total protein N continued to drop. Over time, the percent arginine N in total protein declined from 36 to 5%. This coincided with a 10–33% increase in protein glycine, serine and glutamate N (Table 1). Arginine, released from storage proteins in shoot primordia, now served as an amidino substrate for transamidination reactions.
Radioactivity in 223 and at in the origin of the paper chromatograms were also found as products of [UL-14C]-l-arginine (Durzan 1968). Positive reactions with fast-green and silver nitrate indicated that they were proteinaceous. 14C-Glycine and 14C-serine in acid hydrolysates of these spots would explain their autoradiographic detection. Based on the chromatographic analyses and metabolic profiles in a separate experiment, the traces of 224–226 were not products derived from commercially available 14C-guanidine (unpublished). No evidence was found for the metabolic products which typified with the first three hypotheses.
Discussion
Enzymes responsible for the fate of guanidinoacetic acid
The direct decarboxylation of [1-14C]-guanidinoacetic acid was ruled out because unlabeled N-methylguanidine was not detected on paper chromatograms, autoradiographs, and in the NaOH and NH4OH eluates from the cation-exchange resins on the amino acid analyzer. Second, based on the authentic standards, the methylation of [1-14C]-guanidinoacetic acid to 14C-creatine was ruled out. No evidence was found for 14C-creatine, 14C-creatinine and N-methylglycine (sarcosine) on autoradiographs or in column eluates.
Third, the cleavage of [1-14C]-guanidinoacetic acid by a guanidinoacetase to 14C-glycine and urea was also ruled out. Guanidinoacetases are common in microorganisms (Brunel et al. 1967; Roche et al. 1950; Yorifuji et al. 1977). This study maintained aseptic conditions before and after the study. Guanidinoacetic acid lacking a α-amino group is not used as a substrate by plant arginases. l-Homoarginine and agmatine, having α-amino groups, are substrates for arginase (Dabir et al. 2005; Muszynska and Reifer 1968; Todd et al. 2001).
Transamidination reactions (hypothesis 4) accounted best for the fate of [1-14C]-guanidinoacetic acid to 14C-glycine (Figs. 1, 3). Transamidinases in plants, invertebrates, animals and humans are characterized by their reversibility and wide substrate specificities (Meister 1965; Pisano et al. 1957; Srivenugopal and Adiga 1980; van Thoai 1965). Transamidination systems tend to be conformationally dynamic, functionally promiscuous, contribute to metabolic oscillations, and facilitate the divergence of new functions (Brandman and Meyer 2008, Tokuriki and Tawfik 2009). The transfer of the amidino moiety proceeds without cofactors or ATP.
14C-Glycine was the primary metabolic product in this study (Fig. 3). 14C-Glycine and glycine in the free-amino acid pool were substrates for transamidination. The transamidination of glycine with [1-14C]-guanidinoacetic acid would yield 14C-glycine and guanidinoacetic acid. The transamidination of newly formed 14C-glycine with [1-14C]-guanidinoacetic acid would recycle both [1-14C]-guanidinoacetic acid and 14C-glycine. These amidinotransferases were driven by free glycine, released during the sharp decline of glycine N in shoot storage proteins (Table 1).
High levels of arginine, released by protein turnover, became available as a substrate for an arginine:glycine amidinotransferase. With 14C-glycine, the products of this reaction are [1-14C]-guanidinoacetic acid and ornithine. The transamidination of [1-14C]-guanidinoacetic acid with ornithine yields arginine and 14C-glycine. Arginine may also transfer its amidino moiety to ornithine and regenerate ornithine and arginine as substrates for repeated transamidinations. The results explain how transamidinations accounted for the fate of [1-14C]-guanidinoacetic acid in meristems entering winter dormancy. The destruction of this respiratory inhibitor by transamidination may comprise a mechanism for the awakening from of dormancy and the mobilization of storage protein reserves in shoot primordia.
A wide range of substrates and the reversibility of amidinotransferases are found in animals and plants. Transamidination of [1-14C]-guanidinoacetic acid with glycine to [1-14C]-guanidinoacetic acid and glycine occurs in hog kidneys (Ratner and Rochovansky 1956a, b; Walker 1957). This enzyme also catalyzes transamidinations between canavanine and glycine, and between arginine and canaline. In rat kidney, l-arginine:glycine amidinotransferase transfers the amidino group of l-arginine to ethanolamine, γ-aminobutyrate, δ-aminovalerate, β-aminopropionate, and taurine with the production of ornithine (Watanabe et al. 1994).
In soy bean, l-arginine is a donor of its amidino group to glycine yielding guanidinoacetate and ornithine (Lee et al. 2002). In Lathyrus seedlings, a partial purification of a transamidinase showed a broad pH optimum at 7.5–8.0. It catalyzed the reversible transfer of the amidino group from arginine, homoarginine and canavanine to lysine, putrescine, agmatine, cadaverine and hydroxylysine, respectively (Srivenugopal and Adiga 1980). In Galega officianalis, arginine is a donor of its amidino group for the formation of galegine (3-methyl-but-2-enyl-guanidine) (Reuter 1963; Reuter and Barthel 1967). In jack pine seedlings, transamidination was observed when [amidine-14C]-γ-guanidinobutyric acid and ornithine yielded 14C-arginine and γ-aminobutyric acid as products within 24 h (unpublished).
Transamidination requires that the amidine moiety of a substrate is transferred to a sulfhydryl group on the enzyme to yield a thioamide-enzyme intermediate (Nakatsu 1956; Walker 1957).
Sulfhydryls are strong inhibitors of transamidinase activity (Srivenugopal and Adiga 1980).
The mechanical agitation of shoot primordia in Warburg flasks was sufficient to produce low levels of NO from arginine N and oxygen by a nitric oxide synthase (NOS) (Garcês et al. 2001; Pedroso and Durzan 1999). S-nitrosothiols are formed by the reaction of glutathione or other thiols with nitric oxide (NO) (Durzan and Pedroso 2002; Corpas et al. 2008). NOS activity has been localized in plant peroxisomes (Barroso et al. 1999). S-Nitrosothiolation might protect against the inhibition of sulfhydryls on amidinotransferase activity.
Discussion
Respiration and the oxidative decarboxylation of 14C-glycine and 14C-serine
If ribulose-1,5-bisphosphate (RuBP) was not regenerated as a substrate for O2 or CO2 by photosynthesis and photorespiration, CO2 would be released and trapped. Oxygen would not be needed for the production of glycolate as a substrate for the C2 cycle (photorespiration). In shoot primordia, any photorespiration would be shut down and dark respiration would become a limiting process.
Guanidino compounds are mitochondrial respiratory inhibitors in plants (Wilson and Bonner 1970a, b). Mitochondrial respiration consumes O2 and releases CO2. [1-14C]-guanidinoacetic acid inhibited O2 uptake while the ratios of 14CO2 to 14C-glycine and 14C-serine in the free-amino acid pool slowly increased from 0.2 to 0.6 over 32 h (Table 1). The formation of 14C-glycine and the release of glycine by protein turnover would have enriched their availability to the mitochondrial matrix containing a glycine decarboxylase mitochondrial complex (GDMC). Up to half of all the protein in this matrix comprised the GDMC (Douce et al. 2001; Siedow and Day 2000). Some of 14C-glycine in protein hydrolysates may have included GDMC proteins.
In the GDMC, the P protein would catalyze the formation of a Schiff base between pyridoxal phosphate and 14C-glycine before the oxidative decarboxylation of 14C-glycine to 14CO2. During this process, methylene-H4-folate is used by serine hydroxymethyltransferase to convert 14C-glycine to 14C-serine (Fig. 3). 14C-Serine becomes deaminated to 14C-pyruvate via the TCA cycle (Nelson and Cox 2008).
Ammonium, released by the deamination of 14C-glycine and 14C-serine, would be recovered via α-ketoglutarate from the TCA cycle for the synthesis of traces of 14C-glutamic acid and 14C-proline. The total radioactivity in the organic anion fraction, containing the TCA cycle intermediates and carbohydrates, remained low in shoot primordia (Table 1). In a separate study, 14C-glutamate and 14C-proline, derived from [UL-14C]-L-arginine, are substrates for protein synthesis during bud break in early spring (Durzan 1968, 1969a).
Under the conditions of this study, the mitochondria and peroxisomes are hypothesized as providing arginine-derived NO to maintain organelle function and redox homeostasis in response to the stresses imposed by the respiratory inhibitor [1-14C]-guanidinoacetic acid. The formation of S-nitrosothiols, produced by the reaction of NO and glutathione or other thiols, would protect against the inhibitory activity of sulfhydryls on transamidinase activity. Arginine, NO and guanidino compounds in dormant white spruce have physiological significance for the survival of indigenous peoples in the long winters of eastern Canada (Durzan 2009).
References
Barrenscheen HK, Pany J (1941) Synthetische Leistungen des Keimlings. III. Die Methylierung von Guanidinessigsäure zu Kreatin durch etiolierte Weizenkeimlinge. Biochem Zeitschrift 310:344–349
Barrenscheen HK, von Vály-Nagy T (1942) Die Methylierung durch pflanzliche und tierische Gewebe. I. Methionin als Methylierungsagens bei den Kreatininen und Betainen durch etiolierte Weizenkeimlinge. Hoppe-Seyler’s Zeit für Physiol Chemie 277:97–113
Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupianez JA, del Rio LA (1999) Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem 274:36729–36733
Becker RR (1967) Radiotracer methodology with amino acid analyzers. In: Hirs CHW (ed) Methods in enzymology. Academic Press, New York, pp 108–121
Benson JV, Patterson JA (1965) Accelerated chromatographic analysis of amino acids commonly found in physiological fluids on a spherical resin of specific design. Anal Biochem 37:1108–1110
Brandman O, Meyer T (2008) Feedback loops shape cellular signals in space and time. Science 322:390–395
Brunel A, Brunel-Capelle G, Miquel A (1967) Biosynthèse induite del la l-arginine-uréohydrolase (arginase), de la γ-guanidinobutyrique-uréohydrolase e de la guanidinoacètique-uréohydrolase (glycocyaminase) chez Penicillium roqueforti. Compt Rend Acad Sci Paris 264:2777–2780
Corpas FJ, Chaki M, Frandez-Orcaña A, Valderrama R, Paloma JM, Carreras A, Begara-Morales JC, Airaki M, del Rio LA, Barroso JB (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49:1711–1722
Dabir S, Dabir P, Somvanshi B (2005) Purification, properties and alternate substrate specificities of arginase from two different sources Vigna catjang cotyledon and buffalo liver. Intl J Biol Sci 1:114–122
Douce R, Bourguignon J, Neuburger M, Rébeillè F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6:167–176
Durzan DJ (1968) Nitrogen metabolism of Picea glauca. I. Seasonal changes of free amino acids in buds, shoot apices and leaves, and the metabolism of uniformly labeled 14C-l-arginine by buds during the onset of dormancy. Can J Bot 46:909–919
Durzan DJ (1969a) Nitrogen metabolism of Picea glauca. IV. Metabolism of uniformly labeled 14C-L-arginine, [carbamyl-14C]-L-citrulline, and [1, 2, 3, 4–14C]-γ-guanidinobutyric acid during diurnal changes in the soluble and protein nitrogen associated with the onset of expansion of spruce buds. Can J Biochem 47:771–783
Durzan DJ (1969b) Automated chromatographic analysis of free monosubstituted guanidines in physiological fluids. Can J Biochem 47:657–664
Durzan DJ (2009) Arginine, scurvy, and Jacques Cartier’s “tree of life”. J Ethnobiol Ethnomed 5:5. doi:10.1186/1746-4269-5-5
Durzan DJ, Pedroso MC (2002) Nitric oxide and reactive nitrogen oxide species in plants. Biotech Gen Eng Revs 19:293–337
Garcês H, Durzan DJ, Pedroso MC (2001) Mechanical stress elicits nitric oxide formation and DNA fragmentation in Arabidopsis thaliana. Ann Bot 87:567–574
Lee GT, Kim WJ, Cho YD (2002) Polyamine synthesis in plants. Purification and properties of amindinotransferase from soybean (Glycine max) axes. Phytochemistry 61:781–789
Meister A (1965) Biochemistry of the amino acids, vol II. Academic Press, New York, pp 641–642
Mothes K (1929) Physiologische Untersuchungen uber das Asparagine and das Arginin in Coniferen. Ein Beitrage zur Theorie der Ammoniakentgiftung im pflanzlichen Organismus. Planta 7:585–649
Muszynska G, Reifer I (1968) Purification, properties and inhibition of plant arginase. Acta Biochim Pol 15:55–66
Nakatsu S (1956) Enzymic transamidination from canavanine to glycine by hog kidney extracts. J Biochem (Tokyo) 43:675–683
Nelson DL, Cox MM (2008) Lehninger principles of biochemistry, 5th edn. WH Freeman, San Francisco, pp 863–864
Pedroso MC, Durzan DJ (1999) Detection of apoptosis in chloroplasts and nuclei in different gravitational environments. J Gravit Physiol 6:19–20
Pisano JJ, Mitoma C, Udenfriend S (1957) Biosynthesis of γ-guanidinobutryic acid from γ-aminobutyric acid and arginine. Nature (London) 180:1125–1126
Ratner S, Rochovansky O (1956a) Biosynthesis of guanidinoacetic acid I. Arch Biochim Biophys 63:277–295
Ratner S, Rochovansky O (1956b) Biosynthesis of guanidinoacetic acid. II. Mechanism of amidine group transfer. Arch Biochim Biophys 63:296–315
Reuter G (1963) Arginin als Vorstufe von Galegin in Galega officinalis L. Arch Pharmazie 296:516–522
Reuter G, Barthel A (1967) Guanidinoacetic acid as a precursor of galegine in Galega officinalis L. Pharmazie 22:261
Roche J, LaCombe G, Girard H (1950) On the specificity of certain bacterial deguanidases generating urea and on arginine dihydrolase. Biochim Biophys Acta 6:210–216
Romberger JA (1963) Meristems, growth, and development in woody plants. An analytical review of anatomical, physiological, and morphogenic aspects. USDA Tech Bull No. 1293, pp 214
Schulze E (1896) Über die beim Umsatz der Proteinstoffe in den Keimpflanzen einiger Coniferen Arten entstandenen Stickstoffverbindungen. Hoppe-Seyler’s Zeit für Physiol Chemie 22:435–448
Siedow JN, Day DA (2000) Respiration and photorespiration. Chapter 14. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, MD, pp 676–728
Srivenugopal KS, Adiga PR (1980) Partial purification and properties of a transamidinase from Lathyrus sativus seedlings. Biochem J 189:553–560
Todd CD, Cooke JEK, Gifford DJ (2001) Purification and properties of Pinus taeda arginase from germinated seedlings. Plant Physiol Biochem 39:1037–1045
Tokuriki N, Tawfik DS (2009) Protein dynamism and evolvability. Science 324:203–207
van Thoai N (1965) Nitrogenous bases. In: Florkin M, Stotz EH (eds) Comprehensive biochemistry. Amino Acids and related compounds, vol 5. Amsterdam, Elsevier, pp 208–253
Walker JB (1957) Studies on the mechanism of action of kidney transamidinase. J Biol Chem 224:57–66
Watanabe Y, Van Pilsum JF, Yokoi I, Mori A (1994) Synthesis of neuroactive guanidino compounds by rat kidney l-arginine:glycine amidinotransferase. Life Sci 55:351–358
Weatherley PE, Slatyer RO (1957) Relationship between relative turgidity and diffusion pressure deficit in leaves. Nature (London) 179:1085–1086
Wilson SB, Bonner WD (1970a) Effects of guanidine inhibitors on mung bean mitochondria. Plant Physiol 46:21–24
Wilson SB, Bonner WD (1970b) Energy-linked functions of submitochondrial particles prepared from mung bean mitochondria. Plant Physiol 46:31–35
Woodard JW, Rasch E, Swift H (1961) Nucleic acid and protein metabolism during the mitotic cycle in Vicia faba. J Biophys Biochem Cytol 46:445–462
Wyse M, Kaddurah-Daouk R (2000) Creatine and creatinine metabolism. Physiol Revs 80:1107–1213
Yorifuji T, Tamai H, Usami H (1977) Purification, crystallization and properties of Mn2+ dependent guanidoacetate amidinohydrolase from a Pseudomonas. Agric Biol Chem 41:959–966
Acknowledgments
This work was initially supported by the Canadian Forestry Service at Petawawa on a project with the Chalk River Atomic Energy of Canada on the use of isotopes in tree physiology. The author thanks SM Lopushanski and G Scheer for their help in sampling, scintillation counting and amino acid analyses. Analytic work was completed with McIntyre Stennis funds at University of California at Davis. Studies relating the mechanical agitation of conifer cells to arginine, NO and NOS were supported by a NASA grant (NG 9-825).
Conflict of interest statement
The author declares no conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Rennenberg.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Durzan, D.J. Transamidination of [1-14C]-guanidinoacetic acid to 14C-glycine and decarboxylation to 14CO2 in white spruce shoot primordia entering winter dormancy. Trees 24, 335–341 (2010). https://doi.org/10.1007/s00468-009-0403-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-009-0403-3