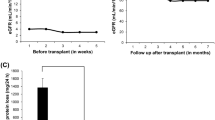
Abstract
Background
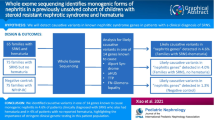
Steroid-resistant nephrotic syndrome (SRNS) is the second most common cause of kidney failure in children and adults under the age of 20 years. Previously, we were able to detect by exome sequencing (ES) a known monogenic cause of SRNS in 25–30% of affected families. However, ES falls short of detecting copy number variants (CNV). Therefore, we hypothesized that causal CNVs could be detected in a large SRNS cohort.
Methods
We performed genome-wide single nucleotide polymorphism (SNP)-based CNV analysis on a cohort of 138 SRNS families, in whom we previously did not identify a genetic cause through ES. We evaluated ES and CNV data for variants in 60 known SRNS genes and in 13 genes in which variants are known to cause a phenocopy of SRNS. We applied previously published, predefined criteria for CNV evaluation.
Results
We detected a novel CNV in two genes in 2 out of 138 families (1.5%). The 9,673 bp homozygous deletion in PLCE1 and the 6,790 bp homozygous deletion in NPHS2 were confirmed across the breakpoints by PCR and Sanger sequencing.
Conclusions
We confirmed that CNV analysis can identify the genetic cause in SRNS families that remained unsolved after ES. Though the rate of detected CNVs is minor, CNV analysis can be used when there are no other genetic causes identified. Causative CNVs are less common in SRNS than in other monogenic kidney diseases, such as congenital anomalies of the kidneys and urinary tract, where the detection rate was 5.3%.
Graphical abstract

A higher resolution version of the Graphical abstract is available as Supplementary information


Similar content being viewed by others
Data availability
Depersonalized data that this study is based on are available from the corresponding author upon request.
References
Barnett HL, Edelmann CM, Greifer I (1981) Primary nephrotic syndrome in children: Clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A report of the international study of kidney disease in children. Kidney Int 20:765–771. https://doi.org/10.1038/ki.1981.209
Leonard MB, Donaldson LA, Ho M, Geary DF (2003) A prospective cohort study of incident maintenance dialysis in children: an NAPRTC study. Kidney Int 63:744–755. https://doi.org/10.1046/j.1523-1755.2003.00788.x
Connaughton DM, Kennedy C, Shril S et al (2019) Monogenic causes of chronic kidney disease in adults. Kidney Int 95:914–928. https://doi.org/10.1016/j.kint.2018.10.031
Wiggins RC (2007) The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int 71:1205–1214
Warejko JK, Tan W, Daga A et al (2018) Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 13:53–62. https://doi.org/10.2215/CJN.04120417
Sadowski CE, Lovric S, Ashraf S et al (2015) A Single-Gene Cause in 29.5% of Cases of Steroid-Resistant Nephrotic Syndrome. J Am Soc Nephrol 26:1279–1289. https://doi.org/10.1681/ASN.2014050489
Mann N, Braun DA, Amann K et al (2019) Whole-Exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol 30:201–215. https://doi.org/10.1681/ASN.2018060575
Zhao L, Liu H, Yuan X et al (2020) Comparative study of whole exome sequencing-based copy number variation detection tools. BMC Bioinformatics 21:97. https://doi.org/10.1186/s12859-020-3421-1
Krumm N, Sudmant PH, Ko A et al (2012) Copy number variation detection and genotyping from exome sequence data. Genome Res 22:1525–1532. https://doi.org/10.1101/gr.138115.112
Klambauer G, Schwarzbauer K, Mayr A et al (2012) Cn.MOPS: Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res 40:e69. https://doi.org/10.1093/nar/gks003
Talevich E, Shain AH, Botton T, Bastian BC (2016) CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 12:e1004873. https://doi.org/10.1371/journal.pcbi.1004873
Sathirapongsasuti JF, Lee H, Horst BAJ et al (2011) Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27:2648–2654. https://doi.org/10.1093/bioinformatics/btr462
Tan R, Wang Y, Kleinstein SE et al (2014) An Evaluation of Copy Number Variation Detection Tools from Whole-Exome Sequencing Data. Hum Mutat 35:899–907. https://doi.org/10.1002/humu.22537
Wu C-HW, Lim TY, Wang C et al (2022) Copy Number Variation Analysis Facilitates Identification of Genetic Causation in Patients with Congenital Anomalies of the Kidney and Urinary Tract. Eur Urol Open Sci 44:106–112. https://doi.org/10.1016/J.EUROS.2022.08.004
Sanna-Cherchi S, Kiryluk K, Burgess KE et al (2012) Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 91:987–997. https://doi.org/10.1016/j.ajhg.2012.10.007
Verbitsky M, Westland R, Perez A et al (2019) The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat Genet 51:117–127. https://doi.org/10.1038/s41588-018-0281-y
Westland R, Verbitsky M, Vukojevic K et al (2015) Copy number variation analysis identifies novel CAKUT candidate genes in children with a solitary functioning kidney. Kidney Int 88:1402–1410. https://doi.org/10.1038/ki.2015.239
Snoek R, van Setten J, Keating BJ et al (2018) NPHP1 (Nephrocystin-1) gene deletions cause adult-onset ESRD. J Am Soc Nephrol 29:1772–1779. https://doi.org/10.1681/ASN.2017111200
Nakanishi K, Okamoto T, Nozu K et al (2019) Pair analysis and custom array CGH can detect a small copy number variation in COQ6 gene. Clin Exp Nephrol 23:669–675. https://doi.org/10.1007/s10157-018-1682-z
Nagano C, Nozu K, Morisada N et al (2018) Detection of copy number variations by pair analysis using next-generation sequencing data in inherited kidney diseases. Clin Exp Nephrol 22:881–888. https://doi.org/10.1007/s10157-018-1534-x
Gee HY, Otto EA, Hurd TW et al (2014) Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85:880–887. https://doi.org/10.1038/ki.2013.450
Hermle T, Schneider R, Schapiro D et al (2018) GAPVD1 and ANKFY1 mutations implicate RAB5 regulation in nephrotic syndrome. J Am Soc Nephrol 29:2123–2138. https://doi.org/10.1681/ASN.2017121312
Hinkes B, Wiggins RC, Gbadegesin R et al (2006) Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 38:1397–1405. https://doi.org/10.1038/ng1918
Boute N, Gribouval O, Roselli S et al (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24:349–354. https://doi.org/10.1038/74166
Swaminathan GJ, Bragin E, Chatzimichali EA et al (2012) DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Hum Mol Genet 21:R37–R44. https://doi.org/10.1093/hmg/dds362
Firth HV, Richards SM, Bevan AP et al (2009) DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 84:524–533. https://doi.org/10.1016/j.ajhg.2009.03.010
Miller DT, Adam MP, Aradhya S et al (2010) Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am J Hum Genet 86:749–764. https://doi.org/10.1016/j.ajhg.2010.04.006
Heeringa SF, Chernin G, Chaki M et al (2011) COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 121:2013–2024. https://doi.org/10.1172/JCI45693
Tan W, Lovric S, Ashraf S et al (2018) Analysis of 24 genes reveals a monogenic cause in 11.1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr Nephrol 33:305–314. https://doi.org/10.1007/s00467-017-3801-6
Av D, Ghosh R, Yuan B et al (2019) Copy number variant and runs of homozygosity detection by microarrays enabled more precise molecular diagnoses in 11,020 clinical exome cases. Genome Med 11:30. https://doi.org/10.1186/s13073-019-0639-5
Boone PM, Campbell IM, Baggett BC et al (2013) Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res 23:1383–1394. https://doi.org/10.1101/gr.156075.113
Lalani SR, Liu P, Rosenfeld JA et al (2016) Recurrent Muscle Weakness with Rhabdomyolysis, Metabolic Crises, and Cardiac Arrhythmia Due to Bi-allelic TANGO2 Mutations. Am J Hum Genet 98:347–357. https://doi.org/10.1016/j.ajhg.2015.12.008
Web resources
ClinVar. National Center for Biotechnology Information, U.S. National Library of Medicine. Accessed July 1, 2023. http://www.ncbi.nlm.nih.gov/clinvar
DECIPHER. Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Accessed July 1, 2023. https://decipher.sanger.ac.uk/
ISCA. International Standards for Cytogenomic Arrays. Accessed July 1, 2023. https://www.clinicalgenome.org/
Acknowledgements
We thank all participating families and physicians for their contribution. F.H. is the William E. Harmon Professor of Pediatrics at Harvard Medical School.
Funding
This research was supported by grants from the Department of Defense to S.S-C. (PR190746 and PR212415), and by grants from the National Institutes of Health to F.H. and S.S-C. (5RC-2DK122397-02), to F.H. (5R01-DK076683-16), and to N.D.M (T5T32-DK007726-37).
Author information
Authors and Affiliations
Contributions
Conceptualization: D.P., F.H., S.S.; Data Curation: D.P., S.S., T.Y.L.; Analysis and interpretation of data: D.P., N.D.M, R.S., S.H., S.S., T.Y.L.; Funding Acquisition: F.H., S.S-C.; Methodology: S.S-C., S.S., T.Y.L.; Project Administration: F.H.; Acquisition of Data: J.A.K., M.A.S., S.E.D.; Software: S.S., T.Y.L.; Supervision: F.H.; Validation: D.P., S.S.; Visualization: D.P.; Writing-original draft: D.P., F.H.; Writing-review and editing: F.H., N.D.M., S.S-C., S.S., T.Y.L.
Corresponding author
Ethics declarations
Ethics approval and consent
This study was approved by the institutional review board of Boston Children’s Hospital as well as institutional review boards of institutions where families were recruited. Before inclusion, written informed consent of each individual or their legal guardians was obtained.
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pantel, D., Mertens, N.D., Schneider, R. et al. Copy number variation analysis in 138 families with steroid-resistant nephrotic syndrome identifies causal homozygous deletions in PLCE1 and NPHS2 in two families. Pediatr Nephrol 39, 455–461 (2024). https://doi.org/10.1007/s00467-023-06134-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00467-023-06134-2