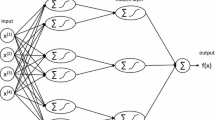
Abstract
We present a probabilistic deep learning methodology that enables the construction of predictive data-driven surrogates for stochastic systems. Leveraging recent advances in variational inference with implicit distributions, we put forth a statistical inference framework that enables the end-to-end training of surrogate models on paired input–output observations that may be stochastic in nature, originate from different information sources of variable fidelity, or be corrupted by complex noise processes. The resulting surrogates can accommodate high-dimensional inputs and outputs and are able to return predictions with quantified uncertainty. The effectiveness our approach is demonstrated through a series of canonical studies, including the regression of noisy data, multi-fidelity modeling of stochastic processes, and uncertainty propagation in high-dimensional dynamical systems.









Similar content being viewed by others
References
Forrester AI, Sóbester A, Keane AJ (2007) Multi-fidelity optimization via surrogate modelling. In: Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences, vol 463. The Royal Society, pp 3251–3269
Robinson T, Eldred M, Willcox K, Haimes R (2008) Surrogate-based optimization using multifidelity models with variable parameterization and corrected space mapping. AIAA J 46:2814–2822
Alexandrov NM, Lewis RM, Gumbert CR, Green LL, Newman PA (2001) Approximation and model management in aerodynamic optimization with variable-fidelity models. J Aircr 38:1093–1101
Sun G, Li G, Stone M, Li Q (2010) A two-stage multi-fidelity optimization procedure for honeycomb-type cellular materials. Comput Mater Sci 49:500–511
Sun G, Li G, Zhou S, Xu W, Yang X, Li Q (2011) Multi-fidelity optimization for sheet metal forming process. Struct Multidiscip Optim 44:111–124
Celik N, Lee S, Vasudevan K, Son Y-J (2010) DDDAS-based multi-fidelity simulation framework for supply chain systems. IIE Trans 42:325–341
Perdikaris P, Karniadakis GE (2016) Model inversion via multi-fidelity Bayesian optimization: a new paradigm for parameter estimation in haemodynamics, and beyond. J R Soc Interface 13:20151107
Perdikaris P (2015) Data-driven parallel scientific computing: multi-fidelity information fusion algorithms and applications to physical and biological systems. Ph.D. thesis, Brown University
Eldred M, Burkardt J (2009) Comparison of non-intrusive polynomial chaos and stochastic collocation methods for uncertainty quantification. In: 47th AIAA aerospace sciences meeting including the new horizons forum and aerospace exposition, p 976
Ng LW-T, Eldred M (2012) Multifidelity uncertainty quantification using non-intrusive polynomial chaos and stochastic collocation. In: 53rd AIAA/ASME/ASCE/AHS/ASC structures, structural dynamics and materials conference 20th AIAA/ASME/AHS adaptive structures conference 14th AIAA, p 1852
Padron AS, Alonso JJ, Palacios F, Barone MF, Eldred MS (2014) Multi-fidelity uncertainty quantification: application to a vertical axis wind turbine under an extreme gust. In: 15th AIAA/ISSMO multidisciplinary analysis and optimization conference, p 3013
Biehler J, Gee MW, Wall WA (2015) Towards efficient uncertainty quantification in complex and large-scale biomechanical problems based on a Bayesian multi-fidelity scheme. Biomech Model Mechanobiol 14:489–513
Peherstorfer B, Willcox K, Gunzburger M (2016) Optimal model management for multifidelity Monte Carlo estimation. SIAM J Sci Comput 38:A3163–A3194
Peherstorfer B, Cui T, Marzouk Y, Willcox K (2016) Multifidelity importance sampling. Comput Methods Appl Mech Eng 300:490–509
Peherstorfer B, Willcox K, Gunzburger M (2016) Survey of multifidelity methods in uncertainty propagation, inference, and optimization. Preprint, pp 1–57
Narayan A, Gittelson C, Xiu D (2014) A stochastic collocation algorithm with multifidelity models. SIAM J Sci Comput 36:A495–A521
Zhu X, Narayan A, Xiu D (2014) Computational aspects of stochastic collocation with multifidelity models. SIAM/ASA J Uncertain Quantif 2:444–463
Bilionis I, Zabaras N, Konomi BA, Lin G (2013) Multi-output separable Gaussian process: towards an efficient, fully Bayesian paradigm for uncertainty quantification. J Comput Phys 241:212–239
Parussini L, Venturi D, Perdikaris P, Karniadakis G (2017) Multi-fidelity Gaussian process regression for prediction of random fields. J Comput Phys 336:36–50
Perdikaris P, Venturi D, Karniadakis GE (2016) Multifidelity information fusion algorithms for high-dimensional systems and massive data sets. SIAM J Sci Comput 38:B521–B538
Rasmussen CE (2004) Gaussian processes in machine learning. In: Bousquet O, von Luxburg U, Rätsch G (eds) Advanced lectures on machine learning. ML 2003. Lecture notes in computer science, vol 3176. Springer, Berlin, Heidelberg, pp 63–71
Kingma DP, Welling, M (2013) Auto-encoding variational Bayes. arXiv preprint arXiv:1312.6114
Sohn K, Lee H, Yan X (2015) Learning structured output representation using deep conditional generative models. In: Cortes C, Lawrence ND, Lee DD, Sugiyama M, Garnett R (eds) Advances in neural information processing systems 28. Curran Associates, Inc., pp 3483–3491
Vincent P, Larochelle H, Bengio Y, Manzagol P-A (2008) Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th international conference on machine learning. ACM, pp 1096–1103
Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol P-A (2010) Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J Mach Learn Res 11:3371–3408
Gómez-Bombarelli R et al (2016) Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat Mater 15:1120–1127
Gómez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, Aspuru-Guzik A (2018) Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci 4:268–276
Ravanbakhsh S, Lanusse F, Mandelbaum R, Schneider JG, Poczos B (2017) Enabling dark energy science with deep generative models of galaxy images. In: AAAI, pp 1488–1494
Lopez R, Regier J, Cole M, Jordan M, Yosef N (2017) A deep generative model for single-cell RNA sequencing with application to detecting differentially expressed genes. arXiv preprint arXiv:1710.05086
Way GP, Greene CS (2017) Extracting a biologically relevant latent space from cancer transcriptomes with variational autoencoders. bioRxiv, pp 174474
Bousquet O, Gelly S, Tolstikhin I, Simon-Gabriel C-J, Schoelkopf B (2017) From optimal transport to generative modeling: the VEGAN cookbook. arXiv preprint arXiv:1705.07642
Pu Y, Chen L, Dai S, Wang W, Li C, Carin L (2017) Symmetric variational autoencoder and connections to adversarial learning. arXiv preprint arXiv:1709.01846
Rosca M, Lakshminarayanan B, Mohamed S (2018) Distribution matching in variational inference. arXiv preprint arXiv:1802.06847
Zheng H, Yao J, Zhang Y, Tsang IW (2018) Degeneration in VAE: in the light of fisher information loss. arXiv preprint arXiv:1802.06677
Kingma DP, Salimans T, Jozefowicz R, Chen X, Sutskever I, Welling M (2016) Improved variational inference with inverse autoregressive flow. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R (eds) Advances in neural information processing systems 29. Curran Associates, Inc., pp 4743–4751
Rezende DJ, Mohamed S (2015) Variational inference with normalizing flows. arXiv preprint arXiv:1505.05770
Burgess CP, Higgins I, Pal A, Matthey L, Watters N, Desjardins G, Lerchner A (2018) Understanding disentangling in \(\beta \)-VAE. arXiv preprint arXiv:1804.03599
Zhao S, Song J, Ermon S (2017) InfoVAE: Information maximizing variational autoencoders. arXiv preprint arXiv:1706.02262
Chen TQ, Li X, Grosse R, Duvenaud D (2018) Isolating sources of disentanglement in variational autoencoders. arXiv preprint arXiv:1802.04942
Burda Y, Grosse R, Salakhutdinov R (2015) Importance weighted autoencoders. arXiv preprint arXiv:1509.00519
Domke J, Sheldon DR (2018) Importance weighting and variational inference. In: Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, Garnett R (eds) Advances in neural information processing systems 31. Curran Associates, Inc., pp 4470–4479
Genevay A, Peyré G, Cuturi M (2017) GAN and VAE from an optimal transport point of view. arXiv preprint arXiv:1706.01807
Villani C (2008) Optimal transport: old and new, vol 338. Springer, Berlin
El Moselhy TA, Marzouk YM (2012) Bayesian inference with optimal maps. J Comput Phys 231:7815–7850
van den Oord A, Kalchbrenner N, Espeholt L, kavukcuoglu k, Vinyals O, Graves Alex (2016) Conditional image generation with PixelCNN decoders. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R (eds) Advances in neural information processing systems 29. Curran Associates, Inc., pp 4790–4798
Liu Q, Wang D (2016) Stein variational gradient descent: a general purpose Bayesian inference algorithm. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R (eds) Advances in neural information processing systems 29. Curran Associates, Inc., pp 2378–2386
Mescheder L, Nowozin S, Geiger A (2017) Adversarial variational bayes: unifying variational autoencoders and generative adversarial networks. arXiv preprint arXiv:1701.04722
Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B (2015) Adversarial autoencoders. arXiv preprint arXiv:1511.05644
Tolstikhin I, Bousquet O, Gelly S, Schoelkopf B (2017) Wasserstein auto-encoders. arXiv preprint arXiv:1711.01558
Titsias MK (2017) Learning model reparametrizations: implicit variational inference by fitting MCMC distributions. arXiv preprint arXiv:1708.01529
Blei DM, Kucukelbir A, McAuliffe JD (2017) Variational inference: a review for statisticians. J Am Stat Assoc 112:859–877
Wainwright MJ, Jordan MI et al (2008) Graphical models, exponential families, and variational inference. Found Trends Mach Learn 1:1–305
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ (eds) Advances in neural information processing systems 27. Curran Associates, Inc., pp 2672–2680
Li C (2018) Towards better representations with deep/Bayesian learning. Duke University
Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X (2016) Improved techniques for training gans. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R (eds) Advances in neural information processing systems 29. Curran Associates, Inc., pp 2234–2242
Akaike H (1998) Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G (eds) Selected papers of Hirotugu Akaike. Springer, Berin, pp 199–213
Friedman J, Hastie T, Tibshirani R (2001) The elements of statistical learning, Springer Series in Statistics, vol 1. Springer, New York
Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, Courville AC (2017) Improved training of Wasserstein GANs. In: Advances in neural information processing systems, pp 5767–5777
Arjovsky M, Chintala S, Bottou L (2017) Wasserstein GAN, arXiv preprint arXiv:1701.07875
Yang L, Zhang D, Karniadakis GE (2018) Physics-informed generative adversarial networks for stochastic differential equations. arXiv preprint arXiv:1811.02033
Schöberl M, Zabaras N, Koutsourelakis P-S (2019) Predictive collective variable discovery with deep Bayesian models. J Chem Phys 150:024109
Grigo C, Koutsourelakis P-S (2019) A physics-aware, probabilistic machine learning framework for coarse-graining high-dimensional systems in the small data regime. arXiv preprint arXiv:1902.03968
Kingma DP, Adam JB (2014) A method for stochastic optimization. arXiv preprint arXiv:1412.6980
Glorot X, Bengio Y (2010) Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp 249–256
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M et al (2016) Tensorflow: a system for large-scale machine learning. In: OSDI, vol 16, pp 265–283
Goodfellow I, Bengio Y, Courville A, Bengio Y (2016) Deep learning, vol 1. MIT Press, Cambridge
Neal RM (2012) Bayesian learning for neural networks, vol 118. Springer, Berlin
Kennedy MC, O’Hagan A (2000) Predicting the output from a complex computer code when fast approximations are available. Biometrika 87:1–13
Perdikaris P, Raissi M, Damianou A, Lawrence N, Karniadakis G (2016) Nonlinear information fusion algorithms for data-efficient multi-fidelity modelling. In: Proceedings of Royal Society A, vol 473. The Royal Society, p 20160751
Burgers JM (1948) A mathematical model illustrating the theory of turbulence. In: von Mises R, von Karman T (eds) Advances in applied mechanics, vol 1. Elsevier, Amsterdam, pp 171–199
Kassam A-K, Trefethen LN (2005) Fourth-order time-stepping for stiff pdes. SIAM J Sci Comput 26:1214–1233
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ (eds) Advances in neural information processing systems 25. Curran Associates, Inc., pp 1097–1105
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Mallat S (2016) Understanding deep convolutional networks. Philos Trans R Soc A 374:20150203
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167
Cohn DA, Ghahramani Z, Jordan MI (1996) Active learning with statistical models. J Artif Intell Res 4:129–145
Shahriari B, Swersky K, Wang Z, Adams RP, De Freitas N (2016) Taking the human out of the loop: a review of Bayesian optimization. Proc IEEE 104:148–175
Yang Y, Perdikaris P (2018) Adversarial uncertainty quantification in physics-informed neural networks. arXiv preprint arXiv:1811.04026
Acknowledgements
This work received support from the US Department of Energy under the Advanced Scientific Computing Research program (Grant DE-SC0019116) and the Defense Advanced Research Projects Agency under the Physics of Artificial Intelligence program (Grant HR00111890034). We would also like to thank the anonymous referees for their constructive feedback.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Sensitivity studies
Appendix: Sensitivity studies
Here we provide results on a series of comprehensive systematic studies that aim to quantify the sensitivity of the resulting predictions on:
-
(i)
the entropic regularization penalty parameter \(\lambda \).
-
(ii)
the generator, discriminator and encoder neural network architectures.
-
(iii)
the adversarial training procedure.
To this end, we consider a simple benchmark corresponding to the approximation of a Gaussian process \(g(x)\sim \mathcal {GP}(\mu _H(x), k(x,x';\theta _H))\), where \(\mu _H(x)\) corresponds to the high-fidelity mean function defined in Eq. 20 and \(k(x,x';\theta _H)\) is a squared exponential kernel with hyper-parameters \(\sigma _{f_H}^2 = 0.5, l_H^2=0.5\), as defined in Eq. 24. Figure 10a shows representative samples generated by this reference stochastic process. In all cases we have employed simple feed-forward neural network architectures as described below. The comparison metric used in all sensitivity studies is the average discrepancy between the predicted and the exact one-dimensional marginal densities, as measured by the reverse Kullback–Leibler divergence
where \(p_1(y|x)\) is the conditional distribution predicted by the generative model, \(p_2(y|x)\) is the conditional distribution of the exact solution, and p(x) is the distribution of uniformly sampled test locations in the interval \(x\in [0,1]\). For a given \(x\sim p(x)\), we facilitate a tractable computation of the reverse KL-divergence using Eq. 25, by performing a Gaussian approximation of \(p_2(y|x)\), while, by definition, \(p_1(y|x)\) is a known uni-variate Gaussian density.
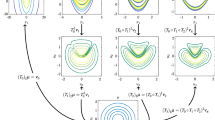
Sensitivity studies on approximating a one-dimensional stochastic process. a Representative samples generated by this reference stochastic process, along with the observed data used for model training. b Representative samples generated by a conditional generative model with \(\lambda =1.5\). Blue lines are the exact reference samples, red markers show the training data, and red lines are the generated samples. (Color figure online)
Sensitivity with respect to the entropic regularization penalty parameter \(\lambda \). a Manifestation of mode collapse for \(\lambda =1.0\). Blue lines are exact samples from the reference stochastic process, red lines are samples produced by the conditional generative model. b Generator and discriminator loss values as a function of the number of training iterations. (Color figure online)
1.1 A.1. Sensitivity with respect to the entropic regularization penalty parameter \(\lambda \)
In this study we aim to quantify the sensitivity of our predictions with respect to the penalty parameter \(\lambda \) in Eq. 10. To this end, we have fixed the architecture for generator and encoder neural networks to include 3 hidden layers with 100 neurons each, and the discriminator neural network to include 2 hidden layers with 100 neurons each. In all cases, we have used a hyperbolic tangent non-linearity and a normal Xavier initialization [64]. For each iteration, we train the discriminator for 3 times and the generator for 1 time. We use a batch size of 500 data-points per stochastic gradient update, and the total number of training points is 10,000.
In Table 1 we report the reverse KL-divergence between the predicted data and the ground truth for different values of \(\lambda \), 1.0, 1.2, 1.5, 1.8, 2.0, and 5.0. Recall that for \(\lambda =1.0\) our model has a direct correspondence with generative adversarial networks [53, 54], while for \(\lambda >1.0\) we obtain a regularized adversarial model that introduces flexibility in mitigating the issue of mode collapse. A manifestation of this pathology is evident in Fig. 11a in which the model with \(\lambda =1.0\) collapses to a degenerate solution that severely underestimates the diversity observed in the true stochastic process samples, despite the fact that the model training dynamics seem to converge to a stable solution (see Fig. 11b). This is also confirmed by the computed average discrepancy in KL-divergence which is roughly an order of magnitude larger compared to the regularized models with \(\lambda >1.0\). We also observe that model predictions remain robust for all values \(\lambda >1.0\), while our best results are typically obtained for \(\lambda =1.5\) which is the value used throughout this paper (see Fig. 10b for representative samples generated by the conditional generative model with \(\lambda =1.5\)).
1.2 A.2. Sensitivity with respect to the neural network architecture
In this study we aim to quantify the sensitivity of our predictions with respect to the architecture of the neural networks that parametrize the generator, the discriminator, and the encoder. Here, we choose the number of layers for the discriminator to always be one less than the number of layers for the generator and the encoder (e.g., if the number of layers for the generator is two then the number of layers for the discriminator is one, etc.). In all cases, we fix \(\lambda = 1.5\) and we use a hyperbolic tangent non-linearity, and a normal Xavier initialization [64]. In Table 2 we report the computed average reverse KL-divergence between the predicted data and the ground truth for different feed-forward architectures for the generator, discriminator, and encoder (i.e., different number of layers and number of nodes in each layer). We denote the number of neurons in each layer as \(N_{n}\) and the number of layers for the generator and the encoder as \(N_g\).
The results of this sensitivity study are summarized in Table 2. Overall, we observe that model predictions remain robust for all neural network architectures considered.
1.3 A.3. Sensitivity with respect to the adversarial training procedure
As discussed in [78], the adversarial training procedure plays a key role in the effectiveness of adversarial generative models, and it often requires a careful tuning of the training dynamics to ensure robustness in the model predictions. To this end, here we test the sensitivity of the proposed conditional generative model with respect to the relative frequency in which the generator and discriminator networks are updated during model training. To this end, we fix we the entropic regularization penalty to \(\lambda = 1.5\), use the neural network architecture to be the same as the one described in “Appendix A.1” section, and vary the total number of training steps for the generator \(K_g\) and the discriminator \(K_d\) within each stochastic gradient descent iteration.
The results of this study are presented in Table 3 where we report the average reverse KL-divergence between the predicted data and the ground truth. These results reveal the high sensitivity of the training dynamics on the interplay between the generator and discriminator networks, and pinpoint the well known peculiarity of adversarial inference procedures which require a careful tuning of \(K_g\) and \(K_d\) for achieving stable performance in practice. Overall we observe that a one-to-three or one-to-five ratio of relative updates for the generator and discriminator, respectively, is the setting that typically works best in practice, although we must underline that this also depends on the capacity of the underlying neural network architectures as discussed in [78].
Finally, Fig. 12 depicts the convergence of the training algorithm for the case \(K_g=1\) and \(K_d=5\). According to [53], the theoretical optimal value of the discriminator loss is \(\ln (4) = - 2 \times \ln (0.5) = 1.384\). As is shown in Fig. 12, the losses oscillate at the very beginning of the training and quickly converge to the optimal value after approximately 2000 iterations.
Optimal convergence of the discriminator loss. Convergence of the training algorithm for the case \(K_g=1\) and \(K_d=5\). The red line depicts the generator loss, the blue line is the discriminator loss, and the black dash line is the theoretical optimal loss of the discriminator. (Color figure online)
Rights and permissions
About this article
Cite this article
Yang, Y., Perdikaris, P. Conditional deep surrogate models for stochastic, high-dimensional, and multi-fidelity systems. Comput Mech 64, 417–434 (2019). https://doi.org/10.1007/s00466-019-01718-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00466-019-01718-y