Abstract
Background
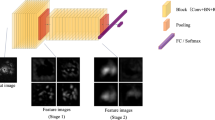
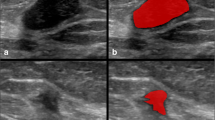
This study explores the application of machine learning (ML) in analyzing endobronchial ultrasound (EBUS) images for the detection of lymph node (LN) malignancy, aiming to augment diagnostic accuracy and efficiency. We investigated whether ML could outperform conventional classification systems in identifying malignant involvement of LNs, based on eight established sonographic features.
Methods
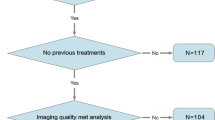
Retrospective data from two tertiary care hospital bronchoscopy units were utilized, encompassing healthcare reports of patients who had undergone EBUS between January 2017 and March 2023. The ML model was trained and tested using MATLAB, with 80% of the data allocated for training/validation, and 20% for testing. Performance was evaluated based on validation and testing accuracy, and receiver operating characteristic curves with comparing trained models and existing classification rules.
Results
The study analyzed 992 LNs, with 42.3% malignancy prevalence. Malignant LNs showed characteristic features such as larger size and distinct margins. The fine tuned models achieved testing accuracies of 95.9% and 96.4% for fine Gaussian SVM and KNN, respectively. Corresponding AUROC’s were 0.955 and 0.963, outperforming other similar studies and conventional analyses.
Conclusion
Fine tuned ML applications like SVM and KNN, can significantly enhance the analysis of EBUS images, improving diagnostic accuracy.


Similar content being viewed by others
References
Alici IO, Yılmaz Demirci N, Yılmaz A, Karakaya J, Özaydın E (2016) The sonographic features of malignant mediastinal lymph nodes and a proposal for an algorithmic approach for sampling during endobronchial ultrasound. Clin Respir J 10:606–613
Hylton DA, Turner S, Kidane B, Spicer J, Xie F, Farrokhyar F, Yasufuku K, Agzarian J, Hanna WC (2020) The Canada Lymph Node Score for prediction of malignancy in mediastinal lymph nodes during endobronchial ultrasound. J Thorac Cardiovasc Surg 159:2499-2507.e2493
Shafiek H, Fiorentino F, Peralta AD, Serra E, Esteban B, Martinez R, Noguera MA, Moyano P, Sala E, Sauleda J, Cosío BG (2014) Real-time prediction of mediastinal lymph node malignancy by endobronchial ultrasound. Arch Bronconeumol 50:228–234
Guberina M, Herrmann K, Pottgen C, Guberina N, Hautzel H, Gauler T, Ploenes T, Umutlu L, Wetter A, Theegarten D, Aigner C, Eberhardt WEE, Metzenmacher M, Wiesweg M, Schuler M, Karpf-Wissel R, Santiago Garcia A, Darwiche K, Stuschke M (2022) Prediction of malignant lymph nodes in NSCLC by machine-learning classifiers using EBUS-TBNA and PET/CT. Sci Rep 12:17511
He L, Huang Y, Yan L, Zheng J, Liang C, Liu Z (2019) Radiomics-based predictive risk score: a scoring system for preoperatively predicting risk of lymph node metastasis in patients with resectable non-small cell lung cancer. Chin J Cancer Res 31:641–652
Laros SSA, Dieckens D, Blazis SP, van der Heide JA (2022) Machine learning classification of mediastinal lymph node metastasis in NSCLC: a multicentre study in a Western European patient population. EJNMMI Phys 9:66
Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, Liu W, Yu L (2017) Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from (18)F-FDG PET/CT images. EJNMMI Res 7:11
Tagaya R, Kurimoto N, Osada H, Kobayashi A (2008) Automatic objective diagnosis of lymph nodal disease by B-mode images from convex-type echobronchoscopy. Chest 133:137–142
Chen CH, Lee YW, Huang YS, Lan WR, Chang RF, Tu CY, Chen CY, Liao WC (2019) Computer-aided diagnosis of endobronchial ultrasound images using convolutional neural network. Comput Methods Programs Biomed 177:175–182
Ozcelik N, Ozcelik AE, Bulbul Y, Oztuna F, Ozlu T (2020) Can artificial intelligence distinguish between malignant and benign mediastinal lymph nodes using sonographic features on EBUS images? Curr Med Res Opin 36:2019–2024
Churchill IF, Gatti AA, Hylton DA, Sullivan KA, Patel YS, Leontiadis GI, Farrokhyar F, Hanna WC (2022) An artificial intelligence algorithm to predict nodal metastasis in lung cancer. Ann Thorac Surg 114:248–256
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10:257–273
Khan S, Yong SP (2016) A comparison of deep learning and hand crafted features in medical image modality classification. In: 2016 3rd international conference on computer and information sciences (ICCOINS), pp 633–638
Funding
This study received no specific grant from any funding agency in the public, commercial or not-for-profit sectors. The authors declare that they have no financial, employment, or other significant/relevant relationships to disclose that could appear to have influenced the submitted work.
Author information
Authors and Affiliations
Contributions
All authors have made substantial contributions to all of the following: (1) the conception and design of the study, or acquisition of data, or analysis and interpretation of data, (2) drafting the article or revising it critically for important intellectual content, (3) final approval of the version to be submitted. All authors also agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The initial draft is authored by the researchers themselves. Subsequently, the English manuscript undergoes refinement and polishing by OpenAI, adhering to native language standards. After using the tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Corresponding author
Ethics declarations
Disclosures
Assistant Professor Fatos Dilan Koseoglu, Professor Ibrahim Onur Alici and Professor Orhan Er have no conflicts of interest or financial ties to disclose.
Ethical approval
This study was approved by the ethical committee of our institution (05042023/969). All patients provided signed informed consent for data acquisition anonymously.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Koseoglu, F.D., Alıcı, I.O. & Er, O. Machine learning approaches in the interpretation of endobronchial ultrasound images: a comparative analysis. Surg Endosc 37, 9339–9346 (2023). https://doi.org/10.1007/s00464-023-10488-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00464-023-10488-x