Abstract
Objective
To investigate the application of machine learning–based ultrasound radiomics in preoperative classification of primary and metastatic liver cancer.
Methods
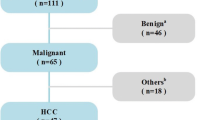
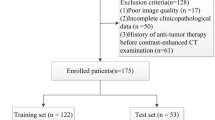
Data of 114 consecutive histopathologically confirmed patients with liver cancer from January 2018 to November 2019 were retrospectively analyzed. All patients underwent liver ultrasonography within 1 week before hepatectomy or fine-needle biopsy. The liver lesions were manually segmented by two experts using ITK-SNAP software. Seven categories of radiomics features, including first-order, two-dimensional shape, gray-level co-occurrence matrices, gray-level run-length matrix, gray-level size-zone matrix, neighboring gray tone difference matrix, and gray-level dependence matrix, were extracted on the Pyradiomics platform. Fourteen filters were applied to the original images, and derived images were obtained. Then, the dimensions of radiomics features were reduced by least absolute shrinkage and selection operator (Lasso) method. Finally, k-nearest neighbor (KNN), logistic regression (LR), multilayer perceptron (MLP), random forest (RF), and support vector machine (SVM) were employed to distinguish primary liver cancer from metastatic liver cancer by a fivefold cross-validation strategy. The performance of the established model was mainly evaluated by the area under the receiver operating characteristic (ROC) curve (AUC) and accuracy.
Results
One thousand four hundred nine radiomics features were extracted from the original images and/or derived images for each patient. The mentioned five machine learning classifiers were able to differentiate primary liver cancer from metastatic liver cancer. LR outperformed other classifiers, with the accuracy of 0.843 ± 0.078 (AUC, 0.816 ± 0.088; sensitivity, 0.768 ± 0.232; specificity, 0.880 ± 0.117).
Conclusions
Machine learning–based ultrasound radiomics features are able to non-invasively distinguish primary liver tumors from metastatic liver tumors.
Key Points
• Ultrasound-based radiomics was initially used for preoperative classification of primary versus metastatic liver cancer.
• Multiple machine learning–based algorithms with cross-validation strategy were applied to extract machine learning–based ultrasound radiomics features.
• Distinction between primary and metastatic tumors was obtained with a sensitivity of 0.768 and a specificity of 0.880.





Similar content being viewed by others
Change history
05 February 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00330-021-07704-4
Abbreviations
- CEUS:
-
Contrast-enhanced ultrasound
- DICOM:
-
Digital Imaging and Communications in Medicine
- FADS:
-
Factor analysis of dynamic structures
- FLL:
-
Focal liver lesion
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run-length matrix
- GLSZM:
-
Gray-level size-zone matrix
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- KNN:
-
k-nearest neighbor
- Lasso:
-
Least absolute shrinkage and selection operator
- LDA:
-
Linear discriminant analysis
- LR:
-
Logistic regression
- MLP:
-
Multilayer perceptron
- NGTDM:
-
Neighboring gray tone difference matrix
- PNN:
-
Probabilistic neural network
- RF:
-
Random forest
- SVM:
-
Support vector machine
- TIC:
-
Time-intensity curve
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Massarweh NN, El-Serag HB (2017) Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Control 24(3):1073274817729245
Bosman FT (2010) WHO classification of tumours of the digestive system, 4th edn. International Agency for Research on Cancer, Lyon
Park JH, Kim JH (2019) Pathologic differential diagnosis of metastatic carcinoma in the liver. Clin Mol Hepatol 25(1):12–20
Liu C-Y, Chen K-F, Chen P-J (2015) Treatment of liver cancer. Cold Spring Harb Perspect Med 5(9):a021535
Koehne de Gonzalez AK, Salomao MA, Lagana SM (2015) Current concepts in the immunohistochemical evaluation of liver tumors. World J Hepatol 7(10):1403–1411
Dagogo-Jack I, Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15(2):81–94
Sumida Y, Nakajima A, Itoh Y (2014) Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol 20(2):475–485
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 281(3):947–957
Mao B, Zhang L, Ning P et al (2020) Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning-based radiomics. Eur Radiol. https://doi.org/10.1007/s00330-020-07056-5
Jin X, Zheng X, Chen D et al (2019) Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur Radiol 29(11):6080–6088
Shiraishi J, Sugimoto K, Moriyasu F, Kamiyama N, Doi K (2008) Computer-aided diagnosis for the classification of focal liver lesions by use of contrast-enhanced ultrasonography. Med Phys 35(5):1734–1746
Rognin NG, Mercier L, Frinking P et al (2009) Parametric imaging of dynamic vascular patterns of focal liver lesions in contrast-enhanced ultrasound. In: 2009 IEEE International Ultrasonics symposium: Rome, Italy September 20–23, 2009. IEEE. Piscataway, NJ, pp 1282–1285
Wu K, Chen X, Ding M (2014) Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik 125(15):4057–4063
Kondo S, Takagi K, Nishida M et al (2017) Computer-aided diagnosis of focal liver lesions using contrast-enhanced ultrasonography with perflubutane microbubbles. IEEE Trans Med Imaging 36(7):1427–1437
Gatos I, Tsantis S, Spiliopoulos S et al (2015) A new automated quantification algorithm for the detection and evaluation of focal liver lesions with contrast-enhanced ultrasound. Med Phys 42(7):3948–3959
Gatos I, Tsantis S, Karamesini M et al (2017) Focal liver lesions segmentation and classification in nonenhanced T2-weighted MRI. Med Phys 44(7):3695–3705
Gatos I, Tsantis S, Karamesini M, Skouroliakou A, Kagadis G (2015) Development of a support vector machine - based image analysis system for focal liver lesions classification in magnetic resonance images. J Phys Conf Ser 633:12116
Parikh T, Drew SJ, Lee VS et al (2008) Focal liver lesion detection and characterization with diffusion-weighted MR imaging: comparison with standard breath-hold T2-weighted imaging. Radiology 246(3):812–822
Mayerhoefer ME, Schima W, Trattnig S, Pinker K, Berger-Kulemann V, Ba-Ssalamah A (2010) Texture-based classification of focal liver lesions on MRI at 3.0 tesla: a feasibility study in cysts and hemangiomas. J Magn Reson Imaging 32(2):352–359
Liang D, Lin L, Hu H et al (2018) Combining convolutional and recurrent neural networks for classification of focal liver lesions in multi-phase CT images. In: Frangi AF (ed) Medical image computing and computer assisted intervention - MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, 2018, proceedings / Alejandro F. Frangi, Julia A. Schnabel, Christos Davatzikos, Carlos Alberola-López, Gabor Fichtinger (eds.). Springer, Cham, pp 666–675
Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet 391(10127):1301–1314
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107
Zwanenburg A, Leger S, Vallières M, Löck S (2016) Image biomarker standardisation initiative. arXiv:1612.07003
Géron A (2017) Hands-on machine learning with Scikit-learn and TensorFlow: concepts, tools, and techniques to build intelligent systems / Aurélien Géron. O'Reilly, Beijing
Zwanenburg A, Leger S, Agolli L et al (2019) Assessing robustness of radiomic features by image perturbation. Sci Rep 9(1):614
Wu M, Li L, Wang J et al (2018) Contrast-enhanced US for characterization of focal liver lesions: a comprehensive meta-analysis. Eur Radiol 28(5):2077–2088
Li W, Huang Y, Zhuang B-W et al (2019) Multiparametric ultrasomics of significant liver fibrosis: a machine learning-based analysis. Eur Radiol 29(3):1496–1506
Yao Z, Dong Y, Wu G et al (2018) Preoperative diagnosis and prediction of hepatocellular carcinoma: radiomics analysis based on multi-modal ultrasound images. BMC Cancer 18(1):1089
Funding
The study was funded by the National Key Research and Development Program of China (Grant No. 2018YFC0114606), National Natural Science Foundation of China (No. 71974065), and Key R & D and Promotion Projects in Henan Province (No. 182400410172).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Lianzhong Zhang.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional review board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The information was missing that Bing Mao and Jingdong Ma contributed equally to this work.
Bing Mao and Jingdong Ma contributed equally to this work.
Supplementary Information
ESM 1
(JPEG 3.12 kb)
Rights and permissions
About this article
Cite this article
Mao, B., Ma, J., Duan, S. et al. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur Radiol 31, 4576–4586 (2021). https://doi.org/10.1007/s00330-020-07562-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07562-6