Abstract
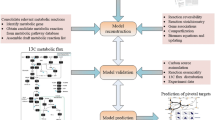
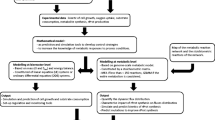
Komagataella phaffii, formerly Pichia pastoris (P. pastoris), is a promising methylotrophic yeast used in industry to produce recombinant protein and valuable metabolites. In this study, a genome-scale metabolic model (GEMs) was reconstructed and used to assess P. pastoris’ metabolic capabilities for the production of S-adenosyl-l-methionine (AdoMet or SAM or SAMe) from individual carbon sources along with the addition of l-methionine. In a model-driven P. pastoris strain, the well-established genome-scale metabolic model iAUKM can be implemented to predict high valuable metabolite production. The model, iAUKM, was created by merging the previously published iMT1026 model and the draught model generated using Raven toolbox from the KEGG database which covered 2309 enzymatic reactions associated with 1033 metabolic genes and 1750 metabolites. The highly curated model was successful in capturing P. pastoris growth on various carbon sources, as well as AdoMet production under various growth conditions. Many overexpression gene targets for increasing AdoMet accumulation in the cell have been predicted for various carbon sources. Inorganic phosphatase (IPP) was one of the predicted overexpression targets as revealed from simulations using iAUKM. When IPP gene was integrated into P. pastoris, we found that AdoMet accumulation increased by 16% and 14% using glucose and glycerol as carbon sources, respectively. Our in silico results shed light on the factors limiting AdoMet production, as well as key pathways for rationalized engineering to increase AdoMet yield.




Similar content being viewed by others
References
Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM (2002) Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 13(4):329–332
Zhan C, Wang S, Sun Y, Dai X, Liu X, Harvey L et al (2016) The Pichia pastoris transmembrane protein GT1 is a glycerol transporter and relieves the repression of glycerol on AOX1 expression. FEMS Yeast Res 16(4):1–10
Zepeda AB, Pessoa A, Farías JG (2018) Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Brazilian J Microbiol 49:119–127. https://doi.org/10.1016/j.bjm.2018.03.010
Bottiglieri T (2002) S-Adenosyl-l-methionine (SAMe): from the bench to the bedside - molecular basis of a pleiotrophic molecule. Am J Clin Nutr 76(5):1151S
Zweier (2014) S-adenosylmethionine metabolism and liver disease. Bone 23(1):1–7
Qin X, Lu J, Zhang Y, Wu X, Qiao X, Wang Z et al (2020) Engineering Pichia pastoris to improve S-adenosyl-l-methionine production using systems metabolic strategies. Biotechnol Bioeng. https://doi.org/10.1002/bit.27300
Gawel LJ, Turner JR, Parks LW (1961) Accumulation of s-adenosylmethionine. 1–3
Shiozaki S, Shimizu S, Yamada H (1984) Unusual intracellular accumulation of s-adenosyl-l-methionine by microorganisms. Agric Biol Chem 48(9):2293–2300
Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y (1988) SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol 8(12):5132–5139
Thomas D, Surdin-Kerjan Y (1991) The synthesis of the two S-adenosyl-methionine synthetases is differently regulated in Saccharomyces cerevisiae. MGG Mol Gen Genet 226(1–2):224–232
Chan SY, Appling DR (2003) Regulation of S-Adenosylmethionine levels in Saccharomyces cerevisiae. J Biol Chem 278(44):43051–43059. https://doi.org/10.1074/jbc.M308696200
Hu H, Qian J, Chu J, Wang Y, Zhuang Y, Zhang S (2009) DNA shuffling of methionine adenosyltransferase gene leads to improved S-adenosyl-l-methionine production in Pichia pastoris. J Biotechnol 141(3–4):97–103
Hu H, Qian J, Chu J, Wang Y, Zhuang Y, Zhang S (2009) Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Appl Microbiol Biotechnol 83(6):1105–1114
Liu W, Tang D, Shi R, Lian J, Huang L, Cai J et al (2019) Efficient production of S-adenosyl-l-methionine from dl-methionine in metabolic engineered Saccharomyces cerevisiae. Biotechnol Bioeng 116(12):3312–3323
Chen Y, Xu D, Fan L, Zhang X, Tan T (2015) Manipulating multi-system of NADPH regulation in Escherichia coli for enhanced S-adenosylmethionine production. RSC Adv 5(51):41103–41111. https://doi.org/10.1039/C5RA02937F
Chen H, Wang Z, Cai H, Zhou C (2016) Progress in the microbial production of S-adenosyl-l-methionine. World J Microbiol Biotechnol 32(9):1–8
Zhao W, Hang B, Zhu X, Wang R, Shen M, Huang L et al (2016) Improving the productivity of S-adenosyl-l-methionine by metabolic engineering in an industrial Saccharomyces cerevisiae strain. J Biotechnol 236:64–70. https://doi.org/10.1016/j.jbiotec.2016.08.003
He J, Deng J, Zheng Y, Gu J (2006) A synergistic effect on the production of S-adenosyl-l-methionine in Pichia pastoris by knocking in of S-adenosyl-l-methionine synthase and knocking out of cystathionine-β synthase. J Biotechnol 126(4):519–527
Hayakawa K, Matsuda F, Shimizu H (2016) Metabolome analysis of Saccharomyces cerevisiae and optimization of culture medium for S-adenosyl-l-methionine production. AMB Express. https://doi.org/10.1186/s13568-016-0210-3
Hu XQ, Chu J, Zhang SL, Zhuang YP, Wang YH, Zhu S et al (2007) A novel feeding strategy during the production phase for enhancing the enzymatic synthesis of S-adenosyl-l-methionine by methylotrophic Pichia pastoris. Enzyme Microb Technol 40(4):669–674
Wang Y, Wang D, Wei G, Wang C (2013) Improved co-production of S-adenosylmethionine and glutathione using citrate as an auxiliary energy substrate. Bioresour Technol 131:28–32. https://doi.org/10.1016/j.biortech.2012.10.168
Ren W, Cai D, Hu S, Xia S, Wang Z, Tan T et al (2017) S-Adenosyl-l-methionine production by Saccharomyces cerevisiae SAM 0801 using DL-methionine mixture: from laboratory to pilot scale. Process Biochem 62(18):48–52. https://doi.org/10.1016/j.procbio.2017.07.014
Ravi Kant H, Balamurali M, Meenakshisundaram S (2014) Enhancing precursors availability in Pichia pastoris for the overproduction of S-adenosyl-l-methionine employing molecular strategies with process tuning. J Biotechnol 188:112–121. https://doi.org/10.1016/j.jbiotec.2014.08.017
Orth JD (2010) Ines Thiele BØP. What is flux balance? Nat Biotechnol 28(3):245–248
Edwards JS, Covert M, Palsson B (2002) Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 4(3):133–140
Wang H, Marcišauskas S, Sánchez BJ, Domenzain I, Hermansson D, Agren R et al (2018) RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol 14(10):1–17
Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J (2013) The RAVEN toolbox and its use for generating a genome-scale metabolic model for penicillium chrysogenum. PLoS Comput Biol 9(3):e100298
Wang W, Kramer PM, Yang S, Pereira MA, Tao L (2001) Reversed-phase high-performance liquid chromatography procedure for the simultaneous determination of S-adenosyl-L-methionine and S-adenosyl-L-homocysteine in mouse liver and the effect of methionine on their concentrations. J Chromatogr B Biomed Sci Appl 762(1):59–65. https://doi.org/10.1016/s0378-4347(01)00341-3
Tomàs-Gamisans M, Ferrer P, Albiol J (2016) Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PLoS ONE 11(1):1–24
Tomàs-Gamisans M, Ferrer P, Albiol J (2018) Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb Biotechnol 11(1):224–237
Ye R, Huang M, Lu H, Qian J, Lin W, Chu J et al (2017) Comprehensive reconstruction and evaluation of Pichia pastoris genome-scale metabolic model that accounts for 1243 ORFs. Bioresour Bioprocess. https://doi.org/10.1186/s40643-017-0152-x
Hartner FS, Glieder A (2006) Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Fact 5:1–21
Sahu U, Rajendra VKH, Kapnoor SS, Bhagavat R, Chandra N, Rangarajan PN (2017) Methionine synthase is localized to the nucleus in Pichia pastoris and Candida albicans and to the cytoplasm in Saccharomyces cerevisiae. J Biol Chem 292(36):14730–14746
Guillamón JM, Van Riel NAW, Giuseppin MLF, Verrips CT (2001) The glutamate synthase (GOGAT) of Saccharomyces cerevisiae plays an important role in central nitrogen metabolism. FEMS Yeast Res 1(3):169–175
Bakker BM, Overkamp KM, Van Maris AJA, Kötter P, Luttik MAH, Van Dijken JP et al (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25(1):15–37
Serrano-Bueno G, Hernández A, López-Lluch G, Pérez-Castiñeira JR, Navas P, Serrano A (2013) Inorganic pyrophosphatase defects lead to cell cycle arrest and autophagic cell death through NAD+ depletion in fermenting yeast. J Biol Chem 288(18):13082–13092
Cregg JM (2007) Pichia protocols. Methods Mol Biol 389:1–10
Acknowledgements
The authors are thankful to Department of Biotechnology (DBT) for providing fellowship to Kabilan S and DBT-Builder Program for providing facility and Dr. Anand Ramaian Santhaseela, Anna University for supporting us.
Funding
This study was supported by Department of Biotechnology, Ministry of Science and Technology, India, BT/PR12153/INF/22/200/2014, BT/PR7605/FNS/20/732/2013.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We affirm that there are no financial, personal, or professional affiliations that might be perceived as influencing the content of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
449_2023_2913_MOESM2_ESM.xml
Supplementary file2 iAUKM model in SBML format. Pichia pastoris GEM model in SBML format generated with the RAVEN toolbox (XML 3397 KB)
449_2023_2913_MOESM3_ESM.xlsx
Supplementary file3 GEMs performance comparison. Analysis of the prediction capabilities of the model iAUKM for identifying the over expression targets for S-adenosyl L-methionine (XLSX 21 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Subash Chandra Bose, K., Shah, M.I., Krishna, J. et al. Genome-scale metabolic model analysis of Pichia pastoris for enhancing the production of S-adenosyl-l-methionine. Bioprocess Biosyst Eng 46, 1471–1482 (2023). https://doi.org/10.1007/s00449-023-02913-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-023-02913-1