Abstract
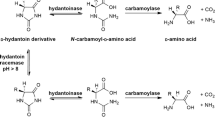
The demand for D-Valine increases because of its wide range of use. A whole-cell biocatalyst for the production of D-Valine from 5′-isopropyl hydantoin by co-expression of the D-hydantoinase (hyd) gene from Pseudomonas putida YZ-26 and D-N-carbamoylase (cab) gene from Sinorhizobium sp. SS-ori in Escherichia coli BL 21 (DE3) was developed. The expression condition of the engineered strain HC01 co-expressing D-hydantoinase (HYD) and D-N-carbamoylase (CAB) was optimized. HYD and CAB reached the highest activities (4.65 and 0.75 U/ml-broth) after inducing for 8–12 h. Subsequently, the cells of HC01 were immobilized in the form of Ca2+-alginate beads, and the optimal conditions for immobilizing were obtained as 2.5% gel concentration and 0.029 g/mL cell concentration in the presence of 3% CaCl2. The thermostability of immobilized cells was 5 ℃ higher than that of free cells in the same condition. And the divalent metal ions such as Mn2+, Mg2+, Cu2+, Co2+, and Ni2+ did not significantly affect the enzymatic activity of HYD and CAB in immobilized cells. Bioconversion rate reached to 91% after a 42-h reaction when the substrate concentration was 50 mmol/L with the initial pH of 9.0 under the nitrogen protection. This method provides D-Valine with optical purity of 97% and an overall yield of 72%. Furthermore, the immobilized cells can be reused for more than 7 cycles and maintain their capacity of over 70%. Hence the immobilized cells of engineered strain HC01 could potentially be used to prepare D-Valine.












Similar content being viewed by others
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Jimenez EC (2020) D-amino acids in peptides from animals, including human: occurrence, structure, bioactivity and pharmacology. Curr Protein Pept Sci 21(6):622–637. https://doi.org/10.2174/1389203721666200426233758
Kapil S, Sharma V (2020) D-amino acids in antimicrobial peptides: a potential approach to treat and combat antimicrobial resistance. Can J Microbiol 67(2):119–137. https://doi.org/10.1139/cjm-2020-0142
Aranaz I, Acosta N, Heras A (2018) Enzymatic D-p-hydrophenyl glycine synthesis using chitin and chitosan as supports for biocatalyst immobilization. Biocatal Biotransform 36(2):1–13. https://doi.org/10.1080/10242422.2017.1366991
Niu L, Shi Y, Yuan J (2004) Fusion expression, purification and enzymatic activity of D-carbamoykase. Chin J Antibiot 29(1):11–14. https://doi.org/10.3969/j.issn.1001-8689.2004.01.004
Clemente-Jiménez JM, Martínez-Rodríguez S, Rodríguez-Vico F, Heras-Vázquez FJ (2008) Optically pure alpha-amino acids production by the “Hydantoinase Process.” Recent Pat Biotechnol 2(1):35–46. https://doi.org/10.2174/187220808783330947
Liu Y, Xu G, Han R, Dong J, Ni Y (2018) Identification of d-carbamoylase for biocatalytic cascade synthesis of d-tryptophan featuring high enantioselectivity. Bioresour Technol 249:720–728. https://doi.org/10.1016/j.biortech.2017.09.162
Nanba H, Ikenaka Y, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci Biotechnol Biochem. 62(5):875–881. https://doi.org/10.1271/bbb.62.875
Ikenaka Y, Nanba H, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Screening, characterization, and cloning of the gene for N-carbamyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci Biotechnol Biochem 62(5):882–886. https://doi.org/10.1271/bbb.62.882
Rodríguez-Alonso MJ, Clemente-Jiménez JM, Rodríguez-Vico F, Las Heras- Vázquez FJ (2015) Rational re-design of the “double-racemase hydantoinase process” for optically pure production of natural and non-natural l-amino acids. Biochem Eng J 101(15):68–76. https://doi.org/10.1016/j.bej.2015.05.003
Pratesi C (1996) Topological mapping of the cysteine residues of N-carbamyl-D-amino-acid amidohydrolase and their role in enzymatic activity. J Biol Chem 271(16):9326–9331. https://doi.org/10.1074/jbc.271.16.9326
Mulla SI, Talwar MP, Hoskeri RS, Ninnekar HZ (2012) Enhanced degradation of 3-nitrobenzoate by immobilized cells of Bacillus flexus strain XJU-4. Biotechnol Bioproc Eng 17(6):1294–1299. https://doi.org/10.1007/s12257-012-0211-2
Tallur PN, Mulla SI, Megadi VB, Talwar MP, Ninnekar HZ (2015) Biodegradation of cypermethrin by immobilized cells of Micrococcus sp. strain CPN 1. Braz J Microbiol 46(3):667–672. https://doi.org/10.1590/S1517-838246320130557
Shi YW, Zhao LX, Niu LX, Xia F, Yuan JM (2005) Gene sequence, soluble expression and homologous comparison of a D-hydantoinase from Pseudomonas putida YZ-26. Chem Res Chin Univ 21(5):552–557
Shi YW, Niu LX, Feng X, Yuan JM (2006) Purification, enzymatic properties of a recombinant d-hydantoinase and its dissociation by zinc ion. World J Microbiol Biotechnol 22(7):675–680. https://doi.org/10.1007/s11274-005-9088-y
Niu LX, Zhang XY, Shi YW, Yuan JM (2007) Subunit dissociation and stability alteration of d-hydantoinase deleted at the terminal amino acid residue. Biotechnol Lett 29(2):303–308. https://doi.org/10.1007/s10529-006-9238-9
Zhang XY, Niu LX, Shi YW, Yuan JM (2008) The flexibility of the non-conservative region at the C terminus of D-hydantoinase from Pseudomonas putida YZ-26 is extremely limited. Appl Biochem Biotechnol 144(3):237–247. https://doi.org/10.1007/s12010-007-8004-3
Zhang XY, Yuan JM, Niu LX, Liang AH (2010) Quantitative analysis and functional evaluation of zinc ion in the d-hydantoinase from Pseudomonas putida YZ-26. Biometals 23(1):71–81. https://doi.org/10.1007/s10534-009-9267-7
Sambrock J, Russel D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Cold Spring Harbor, NY
Nakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T (2000) Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8(7):729–737. https://doi.org/10.1016/s0969-2126(00)00160-x
Aranaz I, Acosta N, Heras A (2018) Enzymatic d-p-hydrophenyl glycine synthesis using chitin and chitosan as supports for biocatalyst immobilization. Biocatal Biotransfor 36(2):89–101. https://doi.org/10.1080/10242422.2017.1366991
Acknowledgements
Supported by Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (STIP) (No. 20110001) and the “1331” project in Shanxi Province.
Funding
The “1331” project in Shanxi Province, scientific and technological innovation programs of higher education institutions in Shanxi (STIP), 20110001.
Author information
Authors and Affiliations
Contributions
LN: Resources, Conceptualization, Writing – review & editing. LX: Formal analysis, Writing – original draft, Visualization. JJ: Formal analysis, Writing – original draft, Investigation. LL: Resources, Conceptualization. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that there are no financial or personal conflicts of interest in publishing this article.
Ethical approval and consent to participate
Not applicable.
Consent for publication
All the authors consent to publish this research paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Niu, L., Xie, L., Ji, J. et al. A novel process using immobilized engineered strain HC01 cells with co-expressed D-hydantoinase and D-N-carbamoylase as biocatalyst for the production of D-Valine. Bioprocess Biosyst Eng 46, 227–236 (2023). https://doi.org/10.1007/s00449-022-02825-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02825-6