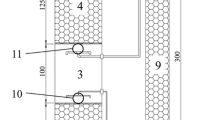
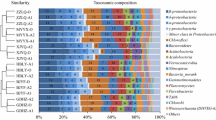
Abstract
Upflow Anaerobic Sludge Blanket (UASB) reactors are alternatives in the anaerobic treatment of sanitary sewage in different parts of the world; however, in temperate environments, they are subject to strong seasonal influence. Understanding the dynamics of the microbial community in these systems is essential to propose operational alternatives, improve projects and increase the quality of treated effluents. In this study, for one year, high-performance sequencing, associated with bioinformatics tools for taxonomic annotation and functional prediction was used to characterize the microbial community present in the sludge of biodigesters on full-scale, treating domestic sewage at ambient temperature. Among the most representative phyla stood out Desulfobacterota (20.21–28.64%), Proteobacteria (7.48–24.90%), Bacteroidota (10.05–18.37%), Caldisericota (9.49–17.20%), and Halobacterota (3.23–6.55%). By performing a Canonical Correspondence Analysis (CCA), Methanolinea was correlated to the efficiency in removing Chemical Oxygen Demand (COD), Bacteroidetes_VadinHA17 to the production of volatile fatty acids (VFAs), and CI75cm.2.12 at temperature. On the other hand, Desulfovibrio, Spirochaetaceae_uncultured, Methanosaeta, Lentimicrobiaceae_unclassified, and ADurb.Bin063-1 were relevant in shaping the microbial community in a co-occurrence network. Diversity analyses showed greater richness and evenness for the colder seasons, possibly, due to the lesser influence of dominant taxa. Among the principal metabolic functions associated with the community, the metabolism of proteins and amino acids stood out (7.74–8.00%), and the genes related to the synthesis of VFAs presented higher relative abundance for the autumn and winter. Despite the differences in diversity and taxonomic composition, no significant changes were observed in the efficiency of the biodigesters.





Similar content being viewed by others
Data availability
The data used to support the findings of this study is available upon request.
References
UNESCO (2019) The United Nations World Water Development Report 2019: Leaving no one behind. UNESCO, Paris
UN-Habitat, WHO (2021) Progress on wastewater treatment: Global status and acceleration needs for SDG indicator 6.3.1. UN Habitat and WHO, Geneva
Sikosana ML, Sikhwivhilu K, Moutloali R, Madyira DM (2019) Municipal wastewater treatment technologies: a review. Procedia Manuf 35:1018–1024. https://doi.org/10.1016/j.promfg.2019.06.051
Abbasi T, Tauseef SM, Abbasi SA (2012) Biogas energy. Biogas Energy. https://doi.org/10.1007/978-1-4614-1040-9
Diamantis V, Eftaxias A, Stamatelatou K et al (2021) Bioenergy in the era of circular economy: anaerobic digestion technological solutions to produce biogas from lipid-rich wastes. Renew Energy 168:438–447. https://doi.org/10.1016/j.renene.2020.12.034
Rehman MLU, Iqbal A, Chang C-C et al (2019) Anaerobic digestion. Water Environ Res 91:1253–1271. https://doi.org/10.1002/wer.1219
Amin FR, Khalid H, El-Mashad HM et al (2021) Functions of bacteria and archaea participating in the bioconversion of organic waste for methane production. Sci Total Environ 763:1–21. https://doi.org/10.1016/j.scitotenv.2020.143007
Mainardis M, Buttazzoni M, Goi D (2020) Up-flow anaerobic sludge blanket (Uasb) technology for energy recovery: a review on state-of-the-art and recent technological advances. Bioengineering 7:1–29. https://doi.org/10.3390/bioengineering7020043
Daud MK, Rizvi H, Akram MF et al (2018) Review of upflow anaerobic sludge blanket reactor technology: effect of different parameters and developments for domestic wastewater treatment. J Chem 2018:1–13. https://doi.org/10.1155/2018/1596319
de Chernicharo CA, Ribeiro TB, Pegorini ES et al (2018) Contribuição para o aprimoramento de projeto, construção e operação de reatores UASB aplicados ao tratamento de esgoto sanitário - Parte 1: Tópicos de Interesse. Rev DAE 66:5–16. https://doi.org/10.4322/dae.2018.038
Cecconet D, Callegari A, Capodaglio AG (2022) UASB performance and perspectives in Urban wastewater treatment at sub-mesophilic operating temperature. Water (Switzerland) 14:1–13. https://doi.org/10.3390/w14010115
Metcalf & Eddy Inc (2014) Wastewater engineering: treatment and resource recovery, 15th edn. McGraw Hill, London
van Lier JB, van der Zee FP, Frijters CTMJ, Ersahin ME (2015) Celebrating 40 years anaerobic sludge bed reactors for industrial wastewater treatment. Rev Environ Sci Bio/Technology 14:681–702. https://doi.org/10.1007/s11157-015-9375-5
ANA (2020) Atualização da Base de Dados de Estações de Tratamento de Esgotos no Brasil. In: ANA (ed) Atlas Esgotos, 1st ed. ANA, Brasília, p. 47
Henze M, Comeau Y (2008) Wastewater Characterization. In: Henze M, van Loosdrecht MCM, Ekama GA, Brdjanovic D (eds) Biological wastewater treatment: principles, modelling and design, 1st edn. IWA Publishing, London, pp 33–53
Zhang B, Yu Q, Yan G et al (2018) Seasonal bacterial community succession in four typical wastewater treatment plants: Correlations between core microbes and process performance. Sci Rep 8:1–11. https://doi.org/10.1038/s41598-018-22683-1
Rashid SS, Liu YQ (2020) Assessing environmental impacts of large centralized wastewater treatment plants with combined or separate sewer systems in dry/wet seasons by using LCA. Environ Sci Pollut Res 27:15674–15690. https://doi.org/10.1007/s11356-020-08038-2
Leitão RC, Van Haandel AC, Zeeman G, Lettinga G (2006) The effects of operational and environmental variations on anaerobic wastewater treatment systems: a review. Bioresour Technol 97:1105–1118. https://doi.org/10.1016/j.biortech.2004.12.007
Marcos A, Al-Kassir A, López F et al (2012) Environmental treatment of slaughterhouse wastes in a continuously stirred anaerobic reactor: Effect of flow rate variation on biogas production. Fuel Process Technol 103:178–182. https://doi.org/10.1016/j.fuproc.2011.12.035
Zhao H, Li J, Li J et al (2013) Organic loading rate shock impact on operation and microbial communities in different anaerobic fixed-bed reactors. Bioresour Technol 140:211–219. https://doi.org/10.1016/j.biortech.2013.04.027
Ketheesan B, Stuckey DC (2015) Effects of hydraulic/organic shock/transient loads in anaerobic wastewater treatment: a review. Crit Rev Environ Sci Technol 45:2693–2727. https://doi.org/10.1080/10643389.2015.1046771
Zhang L, De Vrieze J, Hendrickx TLG et al (2018) Anaerobic treatment of raw domestic wastewater in a UASB-digester at 10 °C and microbial community dynamics. Chem Eng J 334:2088–2097. https://doi.org/10.1016/j.cej.2017.11.073
Callejas C, Fernández A, Passeggi M et al (2019) Microbiota adaptation after an alkaline pH perturbation in a full-scale UASB anaerobic reactor treating dairy wastewater. Bioprocess Biosyst Eng 42:2035–2046. https://doi.org/10.1007/s00449-019-02198-3
Delforno TP, Lacerda Júnior GV, Noronha MF et al (2017) Microbial diversity of a full-scale UASB reactor applied to poultry slaughterhouse wastewater treatment: integration of 16S rRNA gene amplicon and shotgun metagenomic sequencing. Microbiologyopen 6:1–12. https://doi.org/10.1002/mbo3.443
Seib MD, Berg KJ, Zitomer DH (2016) Influent wastewater microbiota and temperature influence anaerobic membrane bioreactor microbial community. Bioresour Technol 216:446–452. https://doi.org/10.1016/j.biortech.2016.05.098
Kuinchtner A, Buriol G (2001) Clima do estado do Rio Grande do Sul segundo a classificação climática de Köppen e Thornthwaite. Discip Sci Série Ciências Exatas 2:171–182
Alvares CA, Stape JL, Sentelhas PC et al (2014) Köppen’s climate classification map for Brazil. Meteorol Zeitschrift 22:711–728. https://doi.org/10.1127/0941-2948/2013/0507
APHA et al (2017) Standard Methods for Examination of Water and Wastewater, 23rd ed. American Public Health Association, Washington
Magrini FE, de Almeida GM, da Maia SD et al (2020) Effect of different heat treatments of inoculum on the production of hydrogen and volatile fatty acids by dark fermentation of sugarcane vinasse. Biomass Convers Biorefinery 11:2443–2456. https://doi.org/10.1007/s13399-020-00687-0
Chernicharo CA de L (2007) Anaerobic Reactors. In: Biological Wastewater Treatment Series, 1st ed. IWA Publishing, London, p. 190
Drosg B, Braun R, Bochmann G, Al Saedi T (2013) Analysis and characterisation of biogas feedstocks. In: Wellinger A, Murphy J, Baxter D (eds) The Biogas Handbook. Elsevier, pp 52–84
INMET (2022) Instituto Nacional de Meteorologia. In: INMET. https://tempo.inmet.gov.br/. Accessed 19 Mar 2022
Caporaso JG, Lauber CL, Walters WA et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Wang Y, Qian PY (2009) Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4:1–9. https://doi.org/10.1371/journal.pone.0007401
Andrews S, Lindenbaum P, Howard B, Ewels P (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. https://doi.org/10.1093/nar/gks1219
Robeson MS, O’Rourke DR, Kaehler BD et al (2021) RESCRIPt: reproducible sequence taxonomy reference database management. PLoS Comput Biol 17:1–37. https://doi.org/10.1371/journal.pcbi.1009581
Whitman WB, Oren A, Chuvochina M et al (2018) Proposal of the suffix –ota to denote phyla. Addendum to ‘proposal to include the rank of phylum in the international code of nomenclature of prokaryotes.’ Int J Syst Evol Microbiol 68:967–969. https://doi.org/10.1099/ijsem.0.002593
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Pielou EC (1966) The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144. https://doi.org/10.1016/0022-5193(66)90013-0
Lozupone C, Lladser ME, Knights D et al (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. https://doi.org/10.1038/ismej.2010.133
Knight R, Vrbanac A, Taylor BC et al (2018) Best practices for analysing microbiomes. Nat Rev Microbiol 16:410–422. https://doi.org/10.1038/s41579-018-0029-9
Douglas GM, Maffei VJ, Zaneveld JR et al (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. https://doi.org/10.1038/s41587-020-0548-6
Czech L, Barbera P, Stamatakis A (2020) Genesis and Gappa: Processing, analyzing and visualizing phylogenetic (placement) data. Bioinformatics 36:3263–3265. https://doi.org/10.1093/bioinformatics/btaa070
Barbera P, Kozlov AM, Czech L et al (2019) EPA-ng: massively parallel evolutionary placement of genetic sequences. Syst Biol 68:365–369. https://doi.org/10.1093/sysbio/syy054
Kanehisa M, Sato Y, Kawashima M et al (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:457–462. https://doi.org/10.1093/nar/gkv1070
Caspi R, Billington R, Ferrer L et al (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:471–480. https://doi.org/10.1093/nar/gkv1164
McNally CP, Eng A, Noecker C et al (2018) BURRITO: an interactive multi-omic tool for visualizing taxa-function relationships in microbiome data. Front Microbiol 9:1–11. https://doi.org/10.3389/fmicb.2018.00365
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Wickham H (2009) ggplot2: elegant graphics for data analysis, 1st edn. Springer, London
Hammer Ø, Harper DAT, Ryan PD (2001) Past: paleontological statistics software package for education and data analysis. Palaentologia Electron 4:1–9
Bastian M, Heymann S, Jacomy M (2009) Gephi: An open source software for exploring and manipulating networks. Int. AAAI Conf. Weblogs Soc. Media 1–2
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. https://doi.org/10.1093/bioinformatics/btu494
IBM Corp. (2011) IBM SPSS Statistics for Windows, Version 20.0
Moyer CL, Eric Collins R, Morita RY (2017) Psychrophiles and psychrotrophs. Ref Modul Life Sci. https://doi.org/10.1016/b978-0-12-809633-8.02282-2
Saavedra O, Escalera R, Heredia G et al (2019) Evaluation of a domestic wastewater treatment plant at an intermediate city in Cochabamba, Bolivia. Water Pract Technol 14:908–920. https://doi.org/10.2166/wpt.2019.071
Lettinga G, Rebac S, Zeeman G (2001) Challenge of psychrophilic anaerobic wastewater treatment. Trends Biotechnol 19:363–370. https://doi.org/10.1016/S0167-7799(01)01701-2
Jordão EP, Pessôa CA (1995) Tratamento de Esgotos Domésticos, 1st edn. ABES, Rio de Janeiro
Von Sperling M (2007) Biological wastewater treatment series: wastewater characteristics, treatment and disposal, 1st edn. IWA Publishing, London
Kang W, Chai H, Xiang Y et al (2017) Assessment of low concentration wastewater treatment operations with dewatered alum sludge-based sequencing batch constructed wetland system. Sci Rep 7:1–7. https://doi.org/10.1038/s41598-017-17783-3
Gaur RZ, Khan AA, Lew B et al (2017) Performance of full-scale uasb reactors treating low or medium strength municipal wastewater. Environ Process 4:137–146. https://doi.org/10.1007/s40710-017-0208-0
da Lobato LC, S, Ribeiro TB, Silva BS da, et al (2018) Contribuição para o aprimoramento de projeto, construção e operação de reatores UASB aplicados ao tratamento de esgoto sanitário - Parte 3: Gerenciamento de lodo e escuma. Rev DAE 66:30–55. https://doi.org/10.4322/dae.2018.040
Bertolino SM, Carvalho CF, Aquino SF (2008) Caracterização e biodegradabilidade aeróbia e anaeróbia dos esgotos produzidos em Campus universitário. Eng sanit Ambient 13:271–277. https://doi.org/10.1590/s1413-41522008000300005
Agrawal LK, Harada H, Okui H (1997) Treatment of dilute wastewater in a UASB reactor at a moderate temperature: Performance aspects. J Ferment Bioeng 83:179–184. https://doi.org/10.1016/S0922-338X(97)83579-9
Alves RGCM, Belli Filho P, Philippi LS, et al (2005) Digestores anaeróbios para tratamento de dejetos suínos: avaliação de partida para diferentes configurações de reatores. In: 23° Congresso Brasileiro de Engenharia Sanitária e Ambiental. ABES, Campo Grande, pp 1–7
Oliveira DBC, Soares WA, Holanda MACR (2020) Effects of rainwater intrusion on an activated sludge sewer treatment system. Rev Ambient e Agua 15:1–12. https://doi.org/10.4136/ambi-agua.2497
Wilén BM, Lumley D, Mattsson A, Mino T (2006) Rain events and their effect on effluent quality studied at a full scale activated sludge treatment plant. Water Sci Technol 54:201–208. https://doi.org/10.2166/wst.2006.721
Tonanzi B, Crognale S, Gianico A et al (2021) Microbial community successional changes in a full-scale mesophilic anaerobic digester from the start-up to the steady-state conditions. Microorganisms 9:1–15. https://doi.org/10.3390/microorganisms9122581
de Lucena RM, Gavazza S, Florencio L et al (2011) Study of the microbial diversity in a full-scale UASB reactor treating domestic wastewater. World J Microbiol Biotechnol 27:2893–2902. https://doi.org/10.1007/s11274-011-0771-x
Calusinska M, Goux X, Fossépré M et al (2018) A year of monitoring 20 mesophilic full-scale bioreactors reveals the existence of stable but different core microbiomes in bio-waste and wastewater anaerobic digestion systems. Biotechnol Biofuels 11:1–19. https://doi.org/10.1186/s13068-018-1195-8
Jiang C, Peces M, Andersen MH et al (2021) Characterizing the growing microorganisms at species level in 46 anaerobic digesters at Danish wastewater treatment plants: a six-year survey on microbial community structure and key drivers. Water Res 193:1–13. https://doi.org/10.1016/j.watres.2021.116871
Liesack W, Bak F, Stackebrandt J-UKE (1994) Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol 162:85–90. https://doi.org/10.1007/s002030050106
Fukunaga Y, Ichikawa N (2014) The Class Holophagaceae. In: Rosenberg E, DeLong EF, Lory S et al (eds) The prokaryotes: other major lineages of bacteria and the archaea, 4th edn. Springer-Verlag, Berlin Heidelberg, pp 683–687
Kielak AM, Barreto CC, Kowalchuk GA et al (2016) The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol 7:1–16. https://doi.org/10.3389/fmicb.2016.00744
Mori K, Yamaguchi K, Sakiyama Y et al (2009) Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. no. Int J Syst Evol Microbiol 59:2894–2898. https://doi.org/10.1099/ijs.0.010033-0
Aida AA, Hatamoto M, Yamamoto M et al (2014) Molecular characterization of anaerobic sulfur-oxidizing microbial communities in up-flow anaerobic sludge blanket reactor treating municipal sewage. J Biosci Bioeng 118:540–545. https://doi.org/10.1016/j.jbiosc.2014.04.011
Waite DW, Chuvochina M, Pelikan C et al (2020) Proposal to reclassify the proteobacterial classes deltaproteobacteria and oligoflexia, and the phylum thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol 70:5972–6016. https://doi.org/10.1099/ijsem.0.004213
Liu Y, Balkwill DL, Henry CA et al (1999) Characterization of the anaerobic propionate- degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int J Syst Bacteriol 49:545–556
Zhang Q, Wang M, Ma X et al (2019) High variations of methanogenic microorganisms drive full-scale anaerobic digestion process. Environ Int 126:543–551. https://doi.org/10.1016/j.envint.2019.03.005
Rinke C, Chuvochina M, Mussig AJ et al (2021) A standardized archaeal taxonomy for the genome taxonomy database. Nat Microbiol 6:946–959. https://doi.org/10.1038/s41564-021-00918-8
Aouad M, Borrel G, Brochier-Armanet C, Gribaldo S (2019) Evolutionary placement of Methanonatronarchaeia. Nat Microbiol 4:558–559. https://doi.org/10.1038/s41564-019-0359-z
Aouad M, Taib N, Oudart A et al (2018) Extreme halophilic archaea derive from two distinct methanogen Class II lineages. Mol Phylogenet Evol 127:46–54. https://doi.org/10.1016/j.ympev.2018.04.011
Laso-Pérez R, Hahn C, van Vliet DM et al (2019) Anaerobic degradation of non-methane alkanes by “ Candidatus Methanoliparia” in hydrocarbon seeps of the Gulf of Mexico. MBio 10:1–20. https://doi.org/10.1128/mBio.01814-19
Rosenberg E, Delong EF, Lory S et al (2014) Other major lineages of bacteria and the archaea, 4th edn. Springer, Berlin
Barton LL, Northup DE (2011) Microbial ecology, 1st edn. Wiley, New Jersey
Taylor HB, Kurtz HD Jr (2019) Composition, diversity, and activity of aerobic ammonia-oxidizing Bacteria and Archaea in the intertidal sands of a grand strand South Carolina beach. Microbiol Open 9:1–18. https://doi.org/10.1002/mbo3.1011
Zhu S, Chen S (2002) The impact of temperature on nitrification rate in fixed film biofilters. Aquac Eng 26:221–237. https://doi.org/10.1016/S0144-8609(02)00022-5
Ju F, Guo F, Ye L et al (2014) Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ Microbiol Rep 6:80–89. https://doi.org/10.1111/1758-2229.12110
Liu F, Hu X, Zhao X et al (2018) Microbial community structures’ response to seasonal variation in a full-scale municipal wastewater treatment plant. Environ Eng Sci. https://doi.org/10.1089/ees.2018.0280
Johnston J, LaPara T, Behrens S (2019) Composition and dynamics of the activated sludge microbiome during seasonal nitrification failure. Sci Rep 9:1–15. https://doi.org/10.1038/s41598-019-40872-4
Zhang Q, Chen X, Luo W et al (2019) Effects of temperature on the characteristics of nitrogen removal and microbial community in post solid-phase denitrification biofilter process. Int J Environ Res Public Health 16:1–15. https://doi.org/10.3390/ijerph16224466
Baldwin SA, Khoshnoodi M, Rezadehbashi M et al (2015) The microbial community of a passive biochemical reactor treating arsenic, zinc, and sulfate-rich seepage. Front Bioeng Biotechnol 3:1–13. https://doi.org/10.3389/fbioe.2015.00027
Chen C, Liang J, Yoza BA et al (2017) Evaluation of an up-flow anaerobic sludge bed (UASB) reactor containing diatomite and maifanite for the improved treatment of petroleum wastewater. Bioresour Technol 243:620–627. https://doi.org/10.1016/j.biortech.2017.06.171
Wang Q, Liang J, Zhan Y et al (2018) Treatment of petroleum wastewater using an up-flow anaerobic sludge blanket (UASB) reactor and turf soil as a support material. J Chem Technol Biotechnol 93:3317–3325. https://doi.org/10.1002/jctb.5694
Mei R, Nobu MK, Narihiro T, Liu WT (2020) Metagenomic and metatranscriptomic analyses revealed uncultured bacteroidales populations as the dominant proteolytic amino acid degraders in anaerobic digesters. Front Microbiol 11:1–11. https://doi.org/10.3389/fmicb.2020.593006
Yamada T, Sekiguchi Y, Hanada S et al (2006) Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the. Int J Syst Evol Microbiol 56:1331–1340. https://doi.org/10.1099/ijs.0.64169-0
Yamada T, Imachi H, Ohashi A et al (2007) Bellilinea caldifistulae gen. nov., sp. nov and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol 57:2299–2306. https://doi.org/10.1099/ijs.0.65098-0
Bovio-Winkler P, Cabezas A, Etchebehere C (2021) Database mining to unravel the ecology of the phylum chloroflexi in methanogenic full scale bioreactors. Front Microbiol 11:1–16. https://doi.org/10.3389/fmicb.2020.603234
Chen S, Liu X, Dong X (2005) Syntrophobacter sulfatireducens sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. Int J Syst Evol Microbiol 55:1319–1324. https://doi.org/10.1099/ijs.0.63565-0
Qiu YL, Hanada S, Ohashi A et al (2008) Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol 74:2051–2058. https://doi.org/10.1128/AEM.02378-07
Imachi H, Sakai S, Sekiguchi Y et al (2008) Methanolinea tarda gen. nov., sp. nov. a methane-producing archaeon isolated from a methanogenic digester sludge. Int J Syst Evol Microbiol 58:294–301. https://doi.org/10.1099/ijs.0.65394-0
Puengrang P, Suraraksa B, Prommeenate P et al (2020) Diverse microbial community profiles of propionate-degrading cultures derived from different sludge sources of anaerobic wastewater treatment plants. Microorganisms 8:1–14. https://doi.org/10.3390/microorganisms8020277
Patel G, Sprott D (1990) Methanosaeta concilii characterization. Int J Syst Bacteriol 40:79–82. https://doi.org/10.1099/00207713-40-1-79
O’Reilly J, Lee C, Collins G et al (2009) Quantitative and qualitative analysis of methanogenic communities in mesophilically and psychrophilically cultivated anaerobic granular biofilims. Water Res 43:3365–3374. https://doi.org/10.1016/j.watres.2009.03.039
Siggins A, Enright AM, O’Flaherty V (2011) Low-temperature (7 °C) anaerobic treatment of a trichloroethylene-contaminated wastewater: Microbial community development. Water Res 45:4035–4046. https://doi.org/10.1016/j.watres.2011.05.013
McKeown RM, Scully C, Enright AM et al (2009) Psychrophilic methanogenic community development during long-term cultivation of anaerobic granular biofilms. ISME J 3:1231–1242. https://doi.org/10.1038/ismej.2009.67
Lee J, Hwang S (2019) Single and combined inhibition of Methanosaeta concilii by ammonia, sodium ion and hydrogen sulfide. Bioresour Technol 281:401–411. https://doi.org/10.1016/j.biortech.2019.02.106
Fernández-Palacios E, Zhou X, Mora M, Gabriel D (2021) Microbial diversity dynamics in a methanogenic-sulfidogenic uasb reactor. Int J Environ Res Public Health 18:1–16. https://doi.org/10.3390/ijerph18031305
Borja R (2011) Biogas production. In: Moo-Young M (ed) Comprehensive biotechnology, 2nd edn. Elsevier, pp 785–798
Hu Y, Jing Z, Sudo Y et al (2015) Effect of influent COD/SO42- ratios on UASB treatment of a synthetic sulfate-containing wastewater. Chemosphere 130:24–33. https://doi.org/10.1016/j.chemosphere.2015.02.019
Wu J, Niu Q, Li L et al (2018) A gradual change between methanogenesis and sulfidogenesis during a long-term UASB treatment of sulfate-rich chemical wastewater. Sci Total Environ 636:168–176. https://doi.org/10.1016/j.scitotenv.2018.04.172
Wu J, Liu Q, Feng B et al (2019) Temperature effects on the methanogenesis enhancement and sulfidogenesis suppression in the UASB treatment of sulfate-rich methanol wastewater. Int Biodeterior Biodegrad 142:182–190. https://doi.org/10.1016/j.ibiod.2019.05.013
Fisher JC, Levican A, Figueras MJ, McLellan SL (2014) Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 5:1–9. https://doi.org/10.3389/fmicb.2014.00525
Rovetto F, Carlier A, Van Den Abeele AM et al (2017) Characterization of the emerging zoonotic pathogen Arcobacter thereius by whole genome sequencing and comparative genomics. PLoS ONE 12:1–27. https://doi.org/10.1371/journal.pone.0180493
Callbeck CM, Pelzer C, Lavik G et al (2019) Arcobacter peruensis sp. nov., a chemolithoheterotroph isolated from sulfide-and organic-rich coastal waters off Peru. Appl Environ Microbiol 85:1–17. https://doi.org/10.1128/AEM.01344-19
Yamada T, Sekiguchi Y (2009) Cultivation of uncultured Chloroflexi subphyla: Significance and ecophysiology of formerly uncultured Chloroflexi “subphylum i” with natural and biotechnological relevance. Microbes Environ 24:205–216. https://doi.org/10.1264/jsme2.ME09151S
Owusu-Agyeman I, Eyice Ö, Cetecioglu Z, Plaza E (2019) The study of structure of anaerobic granules and methane producing pathways of pilot-scale UASB reactors treating municipal wastewater under sub-mesophilic conditions. Bioresour Technol 290:1–9. https://doi.org/10.1016/j.biortech.2019.121733
Liang B, Wang LY, Mbadinga SM et al (2015) Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5:1–13. https://doi.org/10.1186/s13568-015-0117-4
Sun L, Toyonaga M, Ohashi A et al (2016) Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum bacteroidetes, and proposal of lentimicrobiaceae fam. nov. Int J Syst Evol Microbiol 66:2635–2642. https://doi.org/10.1099/ijsem.0.001103
Baena S, Fardeau ML, Labat M et al (1998) Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst Appl Microbiol 21:498–504. https://doi.org/10.1016/S0723-2020(98)80061-1
Lee SH, Park JH, Kang HJ et al (2013) Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresour Technol 145:25–32. https://doi.org/10.1016/j.biortech.2013.02.070
Karami A, Sarshar M, Ranjbar R, Zanjani RS (2014) The Phylum Spirochaetaceae Ali. In: Rosenberg E, De Long EF, Lory S et al (eds) The prokaryotes: other major lineages of bacteria and the archaea. Springer, Berlin, pp 915–925
Delbès C, Moletta R, Godon JJ (2000) Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction- single-strand conformation polymorphism analysis. Environ Microbiol 2:506–515. https://doi.org/10.1046/j.1462-2920.2000.00132.x
Nobu MK, Narihiro T, Rinke C et al (2015) Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9:1710–1722. https://doi.org/10.1038/ismej.2014.256
Probst AJ, Castelle CJ, Singh A et al (2017) Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO 2 concentrations. Environ Microbiol 19:459–474. https://doi.org/10.1111/1462-2920.13362
Martinez MA, Woodcroft BJ, Ignacio Espinoza JC et al (2019) Discovery and ecogenomic context of a global Caldiserica-related phylum active in thawing permafrost, Candidatus Cryosericota phylum nov., Ca. Cryosericia class nov., Ca. Cryosericales ord. nov., Ca. Cryosericaceae fam. nov., comprising the four species C. Syst Appl Microbiol 42:54–66. https://doi.org/10.1016/j.syapm.2018.12.003
Wang D, Huang Q, Wang C et al (2007) The effects of different electron donors on anaerobic nitrogen transformations and denitrification processes in Lake Taihu sediments. Hydrobiologia 581:71–77. https://doi.org/10.1007/s10750-006-0499-z
Dedysh SN, Losey NA, Lawson P (2020) Thermoanaerobaculaceae. In: Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc., pp 1–3
Liu Z, Ma A, Mathé E et al (2021) Network analyses in microbiome based on high-throughput multi-omics data. Brief Bioinform 22:1639–1655. https://doi.org/10.1093/bib/bbaa005
Ma B, Wang H, Dsouza M et al (2016) Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901. https://doi.org/10.1038/ismej.2015.261
Agler MT, Ruhe J, Kroll S et al (2016) Microbial Hub Taxa Link host and abiotic factors to plant microbiome variation. PLoS Biol 14:1–31. https://doi.org/10.1371/journal.pbio.1002352
Berry D, Widder S (2014) Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5:1–14. https://doi.org/10.3389/fmicb.2014.00219
Wang X, Lu X, Li F, Yang G (2014) Effects of temperature and Carbon-Nitrogen (C/N) ratio on the performance of anaerobic co-digestion of dairy manure, chicken manure and rice straw: focusing on ammonia inhibition. PLoS ONE 9:1–7. https://doi.org/10.1371/journal.pone.0097265
Reshmi RR, Deepa Nair K, Zachariah EJ, Vincent SGT (2014) Methanogenesis: seasonal changes in human impacted regions of Ashtamudi estuary (Kerala, South India). Estuar Coast Shelf Sci 156:144–154. https://doi.org/10.1016/j.ecss.2014.11.031
Bandara WMKRTW, Kindaichi T, Satoh H et al (2012) Anaerobic treatment of municipal wastewater at ambient temperature: Analysis of archaeal community structure and recovery of dissolved methane. Water Res 46:5756–5764. https://doi.org/10.1016/j.watres.2012.07.061
Ruiz-Sánchez J, Guivernau M, Fernández B et al (2019) Functional biodiversity and plasticity of methanogenic biomass from a full-scale mesophilic anaerobic digester treating nitrogen-rich agricultural wastes. Sci Total Environ 649:760–769. https://doi.org/10.1016/j.scitotenv.2018.08.165
Pierangeli GMF, Domingues MR, de Jesus TA et al (2021) Higher abundance of sediment methanogens and methanotrophs do not predict the atmospheric methane and carbon dioxide flows in eutrophic tropical freshwater reservoirs. Front Microbiol 12:1–15. https://doi.org/10.3389/fmicb.2021.647921
Sheik CS, Jain S, Dick GJ (2013) Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 16:1–14. https://doi.org/10.1111/1462-2920.12165
Wang HZ, Lv XM, Yi Y et al (2019) Using DNA-based stable isotope probing to reveal novel propionate- and acetate-oxidizing bacteria in propionate-fed mesophilic anaerobic chemostats. Sci Rep 9:1–12. https://doi.org/10.1038/s41598-019-53849-0
Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. https://doi.org/10.1038/nrmicro2259
Paulo LM, Stams AJM, Sousa DZ (2015) Methanogens, sulphate and heavy metals: a complex system. Rev Environ Sci Biotechnol 14:537–553. https://doi.org/10.1007/s11157-015-9387-1
Werner JJ, Knights D, Garcia ML et al (2011) Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci USA 108:4158–4163. https://doi.org/10.1073/pnas.1015676108
Kang X-H, Leng Y, O MM, et al (2020) The seasonal changes of core bacterial community decide sewage purification in sub-plateau municipal sewage treatment plants. Bioprocess Biosyst Eng 43:1609–1617. https://doi.org/10.1007/s00449-020-02352-2
Resende JA, Godon JJ, Bonnafous A et al (2016) Seasonal Variation on microbial community and methane production during anaerobic digestion of cattle manure in Brazil. Microb Ecol 71:735–746. https://doi.org/10.1007/s00248-015-0647-y
Langille MGI, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Reddy GK, Leferink NGH, Umemura M et al (2020) Exploring novel bacterial terpene synthases. PLoS ONE 15:1–20. https://doi.org/10.1371/journal.pone.0232220
Boronat A, Rodríguez-Concepción M (2015) Terpenoid biosynthesis in prokaryotes. In: Schrader J, Bohlmann J (eds) Biotechnology of isoprenoids, 1st edn. Springer, Switzerland, pp 3–18
Ahn S, Jung J, Jang IA et al (2016) Role of glyoxylate shunt in oxidative stress response. J Biol Chem 291:11928–11938. https://doi.org/10.1074/jbc.M115.708149
Huang MH, Li YM, Gu GW (2010) Chemical composition of organic matters in domestic wastewater. Desalination 262:36–42. https://doi.org/10.1016/j.desal.2010.05.037
Rose C, Parker A, Jefferson B, Cartmell E (2015) The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit Rev Environ Sci Technol 45:1827–1879. https://doi.org/10.1080/10643389.2014.1000761
Tian G, Xi J, Yeung M, Ren G (2020) Characteristics and mechanisms of H2S production in anaerobic digestion of food waste. Sci Total Environ 724:137977. https://doi.org/10.1016/j.scitotenv.2020.137977
Zhao C, Dong H, Zhang Y, Li Y (2019) Discovery of potential genes contributing to the biosynthesis of short-chain fatty acids and lactate in gut microbiota from systematic investigation in E. coli. NPJ Biofilms Microbiomes 5:1–8. https://doi.org/10.1038/s41522-019-0092-7
Lovell CR, Leaphart AB (2005) Community-level analysis: Key genes of CO2-reductive acetogenesis. Methods Enzymol 397:454–469. https://doi.org/10.1016/S0076-6879(05)97028-6
Breton-Deval L, Salinas-Peralta I, Aguirre JSA et al (2021) Taxonomic binning approaches and functional characteristics of the microbial community during the anaerobic digestion of hydrolyzed corncob. Energies 14:1–14. https://doi.org/10.3390/en14010066
Yang T, Mbadinga SM, Zhou L et al (2017) Propionate metabolism and diversity of relevant functional genes by in silico analysis and detection in subsurface petroleum reservoirs. World J Microbiol Biotechnol 33:1–10. https://doi.org/10.1007/s11274-017-2350-2
Kircher B, Woltemate S, Gutzki F et al (2022) Predicting butyrate- and propionate-forming bacteria of gut microbiota from sequencing data. bioRxiv. https://doi.org/10.1101/2022.03.06.48315
Hidalgo KJ, Saito T, Silva RS et al (2020) Microbiome taxonomic and functional profiles of two domestic sewage treatment systems. Biodegradation 32:17–36. https://doi.org/10.1007/s10532-020-09921-y
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Funding
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, 001
Author information
Authors and Affiliations
Contributions
JG designed the study, carried out the experiments, analyzed the data, and wrote the paper; NLL carried out the experiments; JI carried out the experiments; FEM supervised the research; SP supervised the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gaio, J., Lora, N.L., Iltchenco, J. et al. Seasonal characterization of the prokaryotic microbiota of full-scale anaerobic UASB reactors treating domestic sewage in southern Brazil. Bioprocess Biosyst Eng 46, 69–87 (2023). https://doi.org/10.1007/s00449-022-02814-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02814-9