Abstract
Recombinant hepatitis B core antigen (HBcAg) molecules, produced in heterologous expression systems, self-assemble into highly homogenous and non-infectious virus-like particles (VLPs) that are under extensive research for biomedical applications. HBcAg production in the methylotrophic yeast P. pastoris has been well documented; however, productivity screening under various residual methanol levels has not been reported for bioreactor processes. HBcAg production under various excess methanol levels of 0.1, 1.0 and 2.0 g L−1 was investigated in this research. Results indicate that, under these particular conditions, the total process and specific protein yields of 876–1308 mg L−1 and 7.9–11.2 mg gDCW−1, respectively, were achieved after 67–75 h of cultivation. Produced HBcAg molecules were efficiently purified and the presence of highly immunogenic, correctly formed and homogenous HBcAg-VLPs with an estimated purity of 90% was confirmed by electron microscopy. The highest reported HBcAg yield of 1308 mg L−1 and 11.2 mg gDCW−1 was achieved under limiting residual methanol concentration, which is about 2.5 times higher than the next highest reported result. A PI-algorithm-based residual methanol concentration feed rate controller was employed to maintain a set residual methanol concentration. Finally, mathematical process models to characterise the vegetative, dead and total cell biomass (Xv, Xd and X), substrate (Glycerol and Methanol) concentration, reactor volume (V), and product (HBcAg) dynamics during cultivation, were identified. A rare attempt to model the residual methanol concentration during induction is also presented.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus core antigen (HBcAg) is a protein subunit of the hepatitis B virus icosahedral core shell. Intracellularly expressed in heterologous production systems such as bacteria, yeasts, plants or mammalian cell cultures, and the recombinant HBcAg molecules self-assemble in to non-infectious virus-like particles (VLPs) [1]. These particles, being highly immunogenic and known to induce significant T-cell responses and high antibody titres in the exposed host organism [2], have been extensively studied and used in vaccine development [3]. Consequently, the efficient production of purified HBcAg-VLPs is a key aspect for both research and commercial applications.
HBcAg-VLPs have been recognised as a promising VLP carrier platform since the 1980s; naturally, they have also been extensively studied [4, 5]. Recent reviews illustrate that HBcAg-VLPs have successfully been employed as epitope carriers in different vaccine candidates against a myriad of diseases, contrary to its fraternal HBV surface antigen (HBsAg), still lack successful commercial examples [3, 6]. A promising HBcAg-VLP-based commercial vaccine candidate against malaria (MalariVax) was in development; however, it was discontinued after failing the sporozoite challenge [7]. On the contrary, a malaria vaccine candidate (Mosquirix™) based on HBsAg-VLPs was recently approved by the WHO for general use in Africa [8]. Several HBcAg-VLP-based vaccine patent applications can also be found against various diseases: melioidosis [9], hepatocarcinoma, and hepatitis B [10, 11], and HPV [12]. Despite the lack of successful commercial HBcAg-VLP-based vaccines, this platform still stands out as one of the frontrunners for future vaccine design.
A wide range of expression systems are available for recombinant protein production and each of them has their own advantages. The choice of a production platform is in large part dictated by the properties of the target protein—its structure and biological activity [13]. For the past 20–30 years, the P. pastoris expression system has been successfully employed for the production of various recombinant proteins for both research and industrial applications. Some of the main characteristics that make this methylotrophic yeast so suited for foreign protein expression include easy genetic manipulation, high-frequency DNA transformation, cloning by functional complementation, high levels of intra- and extracellularly expressed protein, and the ability to perform higher eukaryotic protein modifications (glycosylation, disulphide bond formation, and proteolytic processing) [14]. Also, relatively low levels of native secreted proteins allow for the use of simple purification strategies of the secreted recombinant proteins. When the economic aspects (high cell growth in minimal media and high product stability in prolonged processes) are taken into consideration, together with the powerful genetic techniques available, P. pastoris is clearly the system of choice for heterologous protein expression [15].
The two-stage cultivation on glycerol and methanol associated with Invitrogen’s “Pichia Fermentation Process Guidelines” is well documented and present in most AOX1-promoter-based P. pastoris cultivation strategies. However, recent trends advocate for a move away from standard protocols towards a more conceptual approach, which allows for the development of process-specific strategies tailored both to the specific combination of product/genetic construct and the characteristics of the bioreactor equipment [16]. The standardized approach employs a constant substrate feed rate during methanol induction. Since cell biomass concentration keeps increasing, but the substrate feed rate remains constant, the amount of substrate per biomass (g DCW−1 h−1) gradually decreases, resulting in a decreasing specific growth rate and, consequently, productivity [16].
A myriad of different methanol feeding strategies have been reported with excellent recombinant product yields [16,17,18]. Two distinct groups can be identified—co-feeding and -stat strategies. Co-feeding strategies aim to supplement the methanol feed with additional substrates (glycerol, sorbitol, mannitol, etc.), whereas -stat strategies attempt to control the methanol feed in a way that a cultivation parameter (µ, DO, methanol concentration) remains at a pre-set value. One of the more popular approaches is the methanol-stat strategy, in which the residual methanol concentration is controlled at a set-point value during the methanol induction phase [17]. There are reports of this strategy leading to increased recombinant protein yields for several products; however, no reports can be found on the effect this strategy has on HBcAg production in P. pastoris.
The residual methanol concentration in AOX1-promoter-regulated P. pastoris systems is a critical factor to control, as it directly influences the production and proteolytic degradation of recombinant proteins, cell growth, lysis, and oxygen transfer; thus, an optimal methanol feeding strategy is crucial to maximize product yields [19]. Residual methanol concentrations between 2.0 and 3.5 g L−1 are considered optimal for protein production with P. pastoris [19]; however, some authors report success with concentrations in 4.0–6.0 g L−1 range [20, 21]. Monitoring and controlling the residual methanol concentration in the cultivation broth are crucial; therefore, a suitable methanol sensor and a methanol feed rate control algorithm should be used to ensure adequate methanol concentration control at a selected set-point.
Two types of control systems for residual methanol concentration control can be distinguished—open loop and closed loop. The open-loop systems do not require any on-line measurements (feedback); however, closed loop systems do. In this particular case, the methanol sensor on-line signal can be used as feedback for methanol feed rate control. Additionally, a control algorithm is necessary to successfully control the residual methanol concentration around set-point value with minimal deviation. For certain residual methanol level control, PI- [22] or PID- [23] control algorithms have been successfully employed.
Stringent requirements for producing biopharmaceutical products and their marketing in the corresponding regions, in accordance with good manufacturing practice (GMP) guidelines, are imposed by the EU [24], USA [25], and other developed countries. Since biotechnological microorganism cultivation processes are highly complex and their productivity is sensitive to optimal process parameter control, the above-mentioned directives require that, to ensure a high level of product quality and process repeatability (safety), advanced microorganism culture process control techniques must be employed. It is also mentioned that model-based methods facilitate an in-depth understanding of the process and expedite the performance of the procedures of cultivation process validation and revalidation [26]. Mathematical modelling of recombinant P. pastoris cultivation processes is well described; however, there are few examples modelling residual methanol concentrations during induction [27,28,29,30,31].
In the present work, we establish a P. pastoris cultivation procedure for high-level expression of HBcAg in a defined medium under various residual methanol levels controlled by the PI-feed rate control algorithm. A previously reported purification method [32] was employed with minor modifications, leading to superior results and producing HBcAg-VLPs with an estimated purity of 90%. Mathematical modelling was used to tune the Kp parameter for residual methanol PI-control algorithm. Suitable mathematical models and model parameters to model biomass growth, substrate (glycerol and methanol) consumption, and HBcAg formation were successfully identified.
Materials and methods
Construction of an expression vector and selection of clones
The construction of the expression vector pPIC-HBc and the selection of an HBc producer clone was described in [32].
Experimental conditions
Cultivation processes were carried out using a recombinant Pichia pastoris GS115 Mut+ strain. Batch cultivation and feeding media solutions used in this study were prepared according to the “Pichia fermentation process guidelines” by Invitrogen Corporation [33]: 1.9 l of Basal Salts Medium [26.7 ml L−1 H3PO4 85%, 0.93 g L−1 CaSO4, 18.2 g L−1 K2SO4, 14.9 g L−1 MgSO4·7H2O, 4.13 g L−1 KOH, 40.0 g L−1 glycerol and 4.35 ml L−1 PTM1 trace-element solution (0.02 g L−1 H3BO4, 5 ml L−1 H2SO4 98%, 6.0 g L−1 CuSO4·5H2O, 0.08 g L−1 NaI, 3.0 g L−1 MnSO4·H2O, 0.2 g L−1 Na2MoO4·2H2O, 0.5 g L−1 Ca2SO4·2H2O, 20.0 g L−1 ZnCl2, 65.0 g L−1 FeSO4·7H2O, 0.2 g L−1 biotin)], was inoculated with 100 mL of inoculum grown in BMGY medium (10.0 g L−1 yeast extract, 20.0 g L−1 peptone, 100 mM potassium phosphate buffer, pH 6.0, 13.4 g L−1 yeast nitrogen base, 10.0 g L−1 glycerol, and 0.0004 g L−1 biotin) at 30 °C for 18–22 h in a shake flask at 250 RPM. Two feeding solutions were used—glycerol fed-batch solution (50% glycerol, 12 mL L−1 PTM1) and methanol fed-batch solution (100% methanol, 12 mL L−1 PTM1).
The bioreactor vessel was filled with distilled water and sterilized at 121 °C for 30 min. BSM, BMGY, and glycerol fed-batch solutions were autoclaved separately at 121 °C for 30 min. PTM1 trace element and methanol fed-batch solutions were sterilized by filtration through a 0.2 µm filter.
Fermentations were carried out in a 5 l (2–4 l working volume) fully automated bench-top fermenter (Biotehniskais centrs, EDF-5.4/BIO-4, Latvia) schematically illustrated in Fig. 1. A calibrated pH sensor probe (Hamilton, EasyFerm Bio, Switzerland) was used to measure the medium pH during cultivation. Before the start of the cultivation process, the fermentation medium pH was adjusted at 5.0 ± 0.1 using a 28% NH4OH solution, which was also used to maintain the set pH value during fermentation. The temperature was controlled at 30.0 ± 0.1 °C. The dissolved oxygen level was measured with a calibrated DO probe (Hamilton, OxyFerm, Switzerland) and kept above 30 ± 5% during fermentation by increasing the stirrer speed (200–1000 RPM) or inlet air enrichment with pure O2. Constant air or air/oxygen mixture at a flow rate of 3.0 slpm was maintained during all processes. A condenser was used to condense the moisture from outlet gasses and excessive foam formation was controlled by adding antifoam 204 (Sigma) when necessary. Substrate feed solutions were pumped using a pre-calibrated high-accuracy peristaltic pump (Longer-Pump, BT100–2 J, China).
The cultivations were initiated with a glycerol batch phase. After 18–24 h, all batch glycerol is consumed and glycerol fed-batch solution is fed into the reactor at a rate of 0.61 mL min−1 for 4 h and until an optical density of 100–120 is reached. Then, the feed substrate is switched to methanol and the solution is fed into the reactor at a rate of 0.12 mL min−1 for 5 h, then at a rate of 0.24 mL min−1 for 2 h, and finally at a rate of 0.36 mL min−1 up until 40 h of cultivation. At 40 h, a direct residual methanol concentration control using a PI-algorithm-based feed rate control is initiated at a selected set-point and carried out until the end of the process.
Analytical measurements
Cell growth was observed by off-line measurements of the culture optical density (OD) at a wavelength of 590 nm (GRANAT, KFK-2, St. Petersburg, Russia). Wet cell weight (WCW) and dry cell weight (DCW) measurements were determined gravimetrically. Biomass samples were placed in pre-weighted Eppendorf® tubes and centrifuged at 15,500g for 5 min. Afterwards, the supernatant was discarded, and the cells resuspended in distilled water and centrifuged once more. The liquid phase was discarded and the remaining wet cell biomass weighted. Afterwards, samples were dried at 105 °C until constant weight was reached and the dry cell biomass weighted. The DCW measurement was selected as the most accurate cell biomass assessment with an average error of ± 1.0 g L−1 [34].
Glycerol and methanol concentrations were measured off-line using, respectively, HPLC (Agilent, 1100 HPLC) and GC (Agilent, 6890 N GC).
Protein samples taken during cultivation were analysed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), with a 5% stacking and 15% separating polyacrylamide gel (PAAG), according to standard protocols. To visualize the separated protein bands, the gels were stained with 0.4% Coomassie Brilliant Blue (CBB) G-250 dye.
The level of HBcAg production was also estimated by disrupting 20 optical units of yeast cells by glass beads in 200 µL of 20 mM Tris–HCl, pH 8.0, eight times for 0.5 min. Debris was separated by low-speed centrifugation, and the supernatant was serially diluted for an immunodiffusion assay [35] using a polyclonal rabbit anti-HBcAg antibody.
For electron microscopy, the protein samples were adsorbed on carbon–formvar-coated copper grids and negatively stained with 1% uranyl acetate aqueous solution. The grids were examined with a JEM-1230 electron microscope (JEOL Ltd., Tokyo, Japan) at 100 kV.
Methanol concentration control
Residual methanol concentration was monitored in the exhaust gas and correlated to the concentration in the fermentation medium (Bluesens, BCP-EtOH, Germany). A moving average algorithm (step = 10) was used to decrease signal noise and increase overall quality.
For residual methanol concentration control, a direct substrate feed rate PI-control approach proposed by Cos et al. was implemented [22]. Methanol concentration mass balance equation
Methanol concentration change dynamics
’: deviation variables.
If enough short time intervals are taken into account, methanol concentration and methanol consumption rate variation can be considered non-significant (Eq. 4). The smoothing spline technique is applied for methanol concentration derivative calculation
To maintain a constant methanol (inducer) concentration, control Eq. 4 should be updated with the PI feedback term (Eq. 5), to regulate methanol addition aiming at stabilizing the signal around the set methanol concentration value
The combination between the model and the PI algorithm makes it possible for the control parameters, Kp and τI, to remain constant throughout the cultivation. The Kp used was within the range of 0.02–0.05 [L2 g−1 h−1], and, as it will be demonstrated further, was dependent on selected residual methanol concentration; the constant τI = 10 min was used in the experiments.
Mathematical modelling
The macrokinetic model presented by Ren et al. [28] was employed to model cell biomass (Xv, Xd and X) and glycerol concentrations (SGly) during HBcAg-producing P. pastoris cultivations. A non-monotonically increasing function equation was chosen to model the methanol concentration (SMeOH); and the product accumulation dynamics (PHBcAg) were estimated using a Luedeking–Piret model equation. MATLAB® 2018a was used for mathematical modelling.
Macrokinetic models describing the balances of energy and intracellular substances for both glycerol and methanol uptake phases were constructed in matrix form according to Ren et al. [28].
For the glycerol uptake phase
The respective biomass growth rate (μ) was determined using Eq. 6. To account for the characteristic lag phase in P. pastoris cultivations, the specific glycerol uptake rate (qGlyc) was first described using the metabolic regulator model and afterwards with the well-known Monod model. The switch between the two models was initiated by the minimum function
For the methanol uptake phase
The respective biomass growth rate (μ) values were determined by solving the equation. The specific methanol uptake rate (qMeOH) was calculated according to the non-monotonically increasing function first described by Jackson and Edwards [36]
As recombinant product biosynthesis under the AOX1 promoter takes place only during the methanol induction phase, product accumulation was modelled only during this phase and is described by the following equation:
Finally, a bioreactor model was constructed to estimate the vegetative, dead and total cell biomass (Xv, Xd, and X, respectively), and substrate (S) and product (P) concentrations. A model describing the liquid media volume (V) is also included, as the reactor volume directly influences the concentration of all the aforementioned parameters
Model parameter sensitivity analysis
To evaluate the model parameters that have the most impact on modelling results, a model parameter sensitivity analysis procedure was carried out. The value of one selected parameter was increased by 5%, while others remained constant, and the effect on modelled variables was estimated by evaluating the difference of the new and reference root-mean-square errors (RMSE5% and RMSE0, respectively) using the following formula:
Downstream processing of HBcAg
To compare the outcome of recombinant HBcAg-VLPs produced under different cultivation conditions, a well-established protocol from Freivalds et al. with some modifications was used [32].
Briefly, 4.0 g of frozen yeast cells were first suspended in 16 mL of lysis buffer (20 mM Tris–HCl, 100 mM NaCl, 0.1% Triton X-100, pH 8.0) and disrupted with a French press (4 cycles, 10′000 psi). The supernatant was separated by centrifugation (30 min, 15,500g, at 15 °C) and pH was adjusted to 8.0 using 0.5 M NaOH. The supernatant was then incubated at 65 °C for 1 h and subsequently centrifuged (15 min, 15,500g). Solid ammonium sulfate was added to the solution up to a saturation of 40%, incubated at 4 °C for 30 min, and centrifuged once more (15 min, 15,500g). The precipitate was then dissolved in 2 mL of 20 mM Tris–HCl buffer (pH 8.0) and loaded onto a gel filtration Sepharose 4 Fast Flow (50 mL bed volume) column in 20 mM Tris–HCl pH 8.0, 100 mM NaCl connected to an ÄKTA chromatography system (Amersham Biosciences, UK). The fractions containing HBcAg-VLPs were isolated, pooled, and loaded onto an anion-exchange Fractogel DEAE column (20 mL bed volume) connected to an ÄKTA chromatography system (Amersham Biosciences, UK). The column was preequilibrated with 20 mM Tris–HCl pH 8.0, 100 mM NaCl, and run at 5.0 mL min−1. Column-bound proteins were eluted by a linear gradient with the same buffer containing 1 M NaCl.
Purity and quality of HBc VLPs were assessed by SDS-PAGE and electron microscopy and the concentration established by the Bradford assay method.
Results
HBcAg production
Scientific literature lacks successful examples of bioreactor cultivations at increased residual methanol concentrations for HBcAg production with P. pastoris; however, it is standard practice when producing HBcAg in flasks. Whether induction at increased residual methanol concentrations can improve recombinant HBcAg biosynthesis yields in P. pastoris was the main hypothesis investigated during this research. A series of five experiments was performed and three residual methanol levels during the induction phase were investigated − 0.1 (limiting), 1.0 and 2.0 g L−1. The experiments can be summarized as follows:
-
Exp. 1—according to the Invitrogen protocol; after adaptation to growth on methanol (process 30th hour), constant methanol feed solution addition with the rate of 0.36 mL min−1.
-
Exp. 2—after adaptation to growth on methanol (process 32th hour), constant methanol feed solution addition with the rate of 0.4–0.5 mL min−1.
-
Exp. 3—after adaptation to growth on methanol (process 40th hour), methanol feed rate control according to the PI algorithm, methanol set-point – 1.0 g L−1; Kp = 0.05 L2 g−1 h−1, τI = 10 min.
-
Exp. 4—after adaptation to growth on methanol (process 40th hour), methanol feed rate control according to the PI algorithm, methanol set-point – 2.0 g L−1; Kp = 0.05 L2 g−1 h−1, τI = 10 min.
-
Exp. 5—after adaptation to growth on methanol (process 40th hour), methanol feed rate control according to the PI algorithm, methanol set-point – 2.0 g L−1; Kp = 0.02 L2 g−1 h−1, τI = 10 min.
The biomass growth, methanol concentration, methanol feed rate, and HBcAg accumulation dynamics during these experiments are presented in Fig. 2.
Under various residual methanol levels, differing amounts of HBcAg (858–1308 mg L−1) were recovered after 67–75 h of cultivation (40–45 h of induction). Interestingly, opposite to the reports of increased residual methanol levels promoting other recombinant product biosynthesis, the highest HBcAg concentration of 1308 mg L−1 was produced in Exp. 1, which was performed in accordance to the Invitrogen guidelines and without any residual methanol concentration control. Although the higher cell concentration reached in this experiment attributed to an increased overall process productivity, the specific HBcAg yields per DCW were also higher than in other experiments – 11.2 mg g−1. The higher methanol feed rate (0.4–0.5 mL min−1) and a slightly higher reached cell biomass in Exp. 2 did not amount to an increased final HBcAg concentration or specific yield − 996 mg L−1 and 8.3 mg g−1, respectively.
Cultivation in Exp. 3 was carried out under a controlled residual methanol concentration of 1.0 g L−1. The PI-algorithm-based methanol concentration control performed admirably and was able to maintain the residual methanol concentration at 1.0 g L−1 with an average deviation of ± 0.3 g L−1. After 72 h of cultivation, 1 L of culture was removed to assess how the control would adapt to the new conditions. After a sharp methanol concentration increase, the feed rate values were decreased and increased again when the excess residual methanol was consumed. At approx. 78 h, a sudden methanol concentration spike was noted. It is unclear what exactly caused the cell methanol uptake to suddenly drop as such. Although the methanol feed was automatically stopped, it took about 12 h for the cells to consume the excess methanol. The results from this experiment also indicate that the optimal cultivation length for maximum HBcAg production is around 70–75 h, as an overall yield decrease can be noted the next day (92 h). Yet, a lower biomass concentration (92.0 g L−1) and a lower both total and specific productivity were achieved in this experiment in comparison to Exp. 1 − 876 mg L−1 and 9.6 mg g−1, respectively.
Exp. 4 was performed at a residual methanol concentration of 2.0 g L−1; however, the same control parameter values were used as in Exp. 3 and this led to a very poor methanol concentration control with an average deviation of approx. ± 1.0 g L−1. This illustrates the importance of process-specific control parameter tuning. Consequently, a very poor production was shown by SDS-PAGE analysis and a decision was made not to purify the end culture. This experiment was repeated with more suitable PI-control parameter values (KP = 0.02 L2 g−1 h−1) in Exp. 5 and a much better residual methanol concentration control was achieved − 2.0 ± 0.39 g L−1. Consequently, a much better productivity was achieved − 944 mg L−1 and 7.9 mg g−1. Although this productivity is comparable to the one achieved in Exp. 1 and was achieved in 67 h (instead of 75 h), around 20% more methanol was used, which would impact process overall economic feasibility. Taking this into consideration, Exp. 1 still seemingly has the upper hand production-wise.
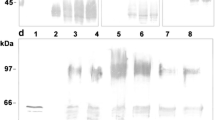
The gel electrophoresis analysis from Exp. 1 in Fig. 3 illustrates the accumulation dynamics of HBcAg during methanol induction. Significant HBcAg accumulation can already be noted in the sample taken just 3.5 h after the start of methanol induction. Despite the thick HBcAg band of the 48th h sample, the specific yield of recombinant protein appears to be approx. 2–3 times higher for the end culture (75th process hour).
Previously processed samples were loaded onto a size-exclusion chromatography column packed with Sepharose 4 Fast flow resin. In laboratory scale, we used a 10 mm Tricorn column, packed at 550 mm with a flow rate of 0.30 mL min−1. HBcAg elution profile is presented in Fig. 4, where the second peak represents the HBcAg VLP fractions (3–7).
SDS-PAGE gel showing HBcAg bands and immunodiffusion titres after each purification step is shown in Fig. 5.
Finally, the collected fractions from the size-exclusion chromatography are merged and loaded onto an anion-exchange column packed with a Fractogel DEAE resin. In laboratory scale, we used a column with a bed volume of 20.0 mL and a flow rate of 4.0 mL min−1. The HBcAg is bound to the column and eluted by a linear salt gradient. The HBcAg elution profile is shown in Fig. 6. Fractions 2 and 3 are collected and pooled for final HBcAg concentration determination and electron microscopy imaging.
The HBcAg concentration in the final fraction was analysed using the universal Bradford assay. It was estimated that the specific HBcAg yield reached 11.2 mg g−1 and the total yield was 1308 mg L−1. The estimated product purity is roughly 90%. The presence of purified, correctly formed, homogenous HBcAg-VLPs is confirmed by the electron microscopy images in Fig. 7. As is apparent from the electron microscopy images, the purified HBcAg-VLPs from samples after 48 h and 75 h of cultivation indicate similar quality.
Mathematical modelling
Along with the Process Analytical Technology (PAT) and Good Manufacturing Practice (GMP) directive-driven requirements for knowledge-based in-depth fermentation process understanding, some typical large-scale culture problems can also be mediated by successfully employing fermentation process mathematical modelling. Namely, to select an appropriate substrate feeding rate profile that provides optimal cell biomass growth and avoids substrate-induced catabolite repression, while also mitigating the very high oxygen demand and metabolic heat production during the methanol induction phase.
When modelling P. pastoris cultivations, the goal should be to refrain from using batch-dependant parameters, and, if possible, to identify comprehensive parameter values that provide suitable accuracy, while remaining constant for all batches. The former approach also sometimes requires time-intensive parameter-identification procedures for every batch, whereas the latter does not.
Here, we present mathematical models to portray cell biomass (Xv, Xd, and X), glycerol (Gly) and methanol (MeOH) concentrations, HBcAg accumulation (P), and reactor volume (V) change dynamics during five HBcAg-producing P. pastoris cultivations. Experimental data were used to evaluate model precision.
To our knowledge, this is the first reported attempt to model HBcAg accumulation during a P. pastoris bioreactor cultivation and the residual methanol concentrations in this particular range. The modelled cultivation experiments are presented in Fig. 8. Identified model parameters and their respective reference values are compiled in Table 1.
Model quality was estimated by means of root-mean-square error (RMSE) and normalized root-mean-square error (NRMSE) values. Calculated values are presented in Table 2.
The model parameter sensitivity analysis results are presented in Table 3. Table values indicate the deviation in each respective modelled variable RMSE, caused by a 5% increase of said model parameter.
The results from the model parameter sensitivity analysis procedure indicate that both glycerol and methanol modelled concentrations are very sensitive to most model parameter values. This can be explained by the relatively low residual concentrations of the substrates in the medium. As for the parameters that have the most impact on modelling results, constants k1 and k2 jump out. Such high sensitivity can be explained, first of all, because they directly influence the biomass (X) concentration, which in turn has a major effect on most other modelled variables. Second, because minor deviations from the biomass growth profile in the early cultivation hours can have a snowball effect in the late process hours.
Discussion
Historically, HBcAg-VLPs have played a key role in vaccine development and, consequently, the efficient production and purification of these particles can play a major role in commercial vaccine production. There are many reports of successfully producing these particles in various recombinant expression platforms; however, P. pastoris stands out due to its ability to reach high cell densities on minimal media, perform post-translational modifications, and drive high-level expression of recombinant proteins. The substrate feeding strategy during the methanol induction phase for recombinant product biosynthesis is recognised to have a significant influence on the product yield and process productivity. Despite this, the effect of residual methanol concentration on HBcAg production with P. pastoris in bioreactor cultivations has not been extensively studied and reported. In this particular research, three residual methanol levels (0.1, 1.0, and 2.0 g L−1) during the HBcAg-producing P. pastoris cultivation methanol induction phase were investigated.
Excellent recombinant HBcAg yields were achieved in all cultivations; however, the best yield was produced in Exp. 1—according to the Invitrogen guidelines and without residual methanol concentration control during induction (1308 mg L−1 and 11.2 mg gDCW−1). When the residual methanol concentration was controlled at 1.0 or 2.0 g L−1, lower HBcAg yields were registered – 876 mg L−1 and 9.6 mg gDCW−1 in Exp. 3, and 944 mg L−1 and 7.8 mg gDCW−1 in Exp. 5, respectively. Additionally, approx. 10–20% more methanol was used in Exps. 3 and 5, which should also be taken into account when evaluating the economic feasibility of a production process. Also, residual methanol concentration control requires both the technical possibility and the implementation and tuning of a suitable methanol feed rate control algorithm. Overall, it is evident that in this particular case, residual methanol concentration control is not feasible. A review of reported P. pastoris-produced HBcAg yields is presented in Table 4.
Unfortunately, experimental screening at higher residual methanol levels (4.0–6.0 g L−1) was not possible, due to the insufficiently low temperature of the tap water used for cooling. Consequently, the bioreactor’s cooling system was unable to successfully maintain the temperature below 32 °C. To avoid this setback, an additional cooling system should be employed, for example, a circulation chiller. Another possible option to avoid cooling issues would be to grow the cells to lower densities; however, this would also lead to lower HBcAg yields. An example can be seen in Exp. 3—where lower cell densities lead to lower overall HBcAg yields, despite the second highest specific HBcAg yield.
The overall HBcAg yield in Exp. 1 is the highest reported in both scientific and patent literature to our knowledge. The second highest reported yield by Freivalds et al. [32] utilizes the same HBc producer clone and a similar purification procedure. There are two main differences. First, BMGY was selected as the cultivation medium. In general, production in minimal (salt) medium, such as BSM, is preferred over rich (complex) BMGY medium, as it eases subsequent product purification and improves overall process economic feasibility. Also, the ability to reach high cell densities and high-level production in minimal medium is one of the main advantages the yeast P. pastoris presents. Second, the authors did not employ a glycerol fed-batch phase, but added a glycerol–methanol mixture to facilitate faster adaptation to methanol consumption. While this may indeed facilitate better cell adaptation to methanol consumption, the absence of a glycerol fed-batch phase leads to lower cell biomass concentrations and, consequently, overall process yields. Although the specific HBcAg yields are comparable (11.2 vs 10.0 mg gDCW−1), we reached an almost 2.5 times higher biomass concentration and, consequently, a 2.5 times higher overall HBcAg yield.
The residual methanol concentration in Exps. 3–5 was controlled by altering the methanol feed rate according to the values calculated by the PI algorithm. This control solution proved to be able to control the residual methanol concentration at set-point value with sufficient accuracy. The main advantage being relatively easy implementation. This controller, however, is quite sensitive to the selected parameter values, as can be seen in Exps. 4 and 5; therefore, a procedure for optimum control parameter value identification is necessary. The control was able to adapt to unexpected situations—for example, when approx. 1 L of culture was removed during Exp. 3 (71 h)—although a brief spike in the residual methanol concentration was noted, it was corrected shortly after. At times, however, the control was prone to increasing oscillations and operator involvement was necessary to stabilize the control signal. Overall, the control proved capable to control the methanol concentration at the set-point value with sufficient accuracy and can be employed in P. pastoris cultivation processes.
The purification procedure used in this research allowed for a rapid and efficient HBcAg VLP purification. When purifying recombinant proteins, an important aspect to consider is the time required for the purification procedure. In this case, the whole procedure can be carried out in 1 day. By following this purification protocol, highly immunogenic, correctly formed and homogenous HBcAg-VLPs with an estimated purity of 90% can be achieved.
As indicated earlier, process mathematical modelling can play an important role in recombinant product development and manufacture and, although there are several reports of successful P. pastoris cultivation process models, there is little information on modelling the residual methanol concentration during the induction phase. In this particular research, we present a process model that characterises cell biomass (Xv, Xd and X), substrate (Gly. and MeOH) concentration, reactor volume (V), and product (HBcAg) accumulation dynamics during cultivation. The experimental data accumulated during these cultivations were used to estimate the model accuracy.
Good results were achieved when modelling the cell biomass (± 6.05 g L−1) and glycerol (± 1.65 g L−1) concentrations. In prior research, using a dielectric spectroscopy probe and an off-gas analyser, we have observed that, during the start of the methanol induction phase—when significant methanol accumulation in the reactor takes place—a reduction in overall cell viability can be noted [34]. The experimental dry cell biomass measurements account also for dead (non-vegetative) cells; therefore, separate models were introduced to account for both vegetative cell (used in substrate and product calculations) and total cell (used for correlation with DCW measurements) biomass concentration. To estimate the dead cell fraction and offset the effect of these non-viable cells present on modelled cell biomass values, an empirical parameter Kd was introduced. This allowed to significantly improve the accuracy of cell biomass estimation, especially in the late induction phase, when the dead cell fraction composes a significant part of total cell biomass. However, some underestimation can still be noted. This is likely because the term Kd does not remain constant during the whole cultivation and is influenced by both the residual methanol concentration and cell exposure (induction) time.
The residual methanol concentration, however, proved difficult to model. Although the model was able to successfully account for methanol concentration peaks at the start of residual methanol concentration control, the overall accuracy could still use more improvement (± 1.26 g L−1). The model also somewhat underestimated the residual methanol concentration during cultivations. Looking at other residual methanol concentration modelling results, the model presented by Ren et al. was unable to account for a fast accumulation of methanol up to 10.0 g L−1 [28]. The model, however, was able to accurately estimate the methanol concentration in a methanol-limited cultivation. Although the model presented by Jahic et al. included methanol concentration estimation, they opted to represent the methanol concentration using experimental measurements, perhaps, thus, indicating that the model was not able to accurately estimate the accumulated methanol concentration up to nearly 2.0 g L−1 during methanol adaptation [27]. A more sophisticated approach is presented by Niu et al., as their model was able to account for methanol accumulation during initial adaptation (approx. 6.4 g L−1) and at the end of the cultivation (approx. 16 g L−1) [29]. Although the model is successful at estimating the residual methanol concentration at the start and the end of the methanol induction, it seemingly fails to account for a small methanol spike in the middle of cultivation. While this presented approach seems promising, it is also complex, as it incorporates real-time exhaust gas analysis and a dynamic parameter-identification procedure, which lessens the potential for practical application. Even though the accuracy of our presented model could still be improved, it demonstrates a rare attempt to model residual methanol concentration with sufficient accuracy and low complexity. The ability of the model to accurately predict increases (spikes) in methanol concentration can be very useful during both laboratory and industrial cultivations, as rapid detection of such spikes may help avoid deviating from the optimal growth trajectory or whole batch discard in general.
Recombinant HBcAg accumulation dynamics were also modelled (± 112.1 mg L−1) and follow the experimentally determined yield values. However, the model could not account for the high productivity in Exp. 1, which could indicate that some other unknown factor could possibly have influenced the high yield in this particular experiment. A significant factor to note is also the fact that this measurement is very dependent on the estimated cell biomass concentration. Ensuring a higher cell biomass estimation precision would also positively impact this measurement in this case.
Overall, we have presented a relatively simple and robust bioreactor model to successfully portray cell biomass (Xv, Xd, and X) concentration, substrate (Gly. and MeOH), reactor volume (V), and product (HBcAg) dynamics during a P. pastoris GS115 cultivation under both limiting (0.1 g L−1) and residual (1.0 and 2.0 g L−1) methanol concentrations. Relatively easy implementation and parameter identification enables this approach to also be suitable for practical applications. The presented model could also be further used for model-based optimization and software-sensor development of the investigated process.
Conclusions
In this particular research, three residual methanol levels (0.1, 1.0, and 2.0 g L−1) during the HBcAg-producing P. pastoris cultivation methanol induction phase were investigated. Increased residual methanol concentration did not have a profound effect on HBcAg accumulation dynamics. Considering the economic aspects of the production process, bioreactor cultivation under limiting methanol concentration, according to the Invitrogen cultivation guidelines, is preferable. The highest reported HBcAg VLP yield of 1308 mg L−1 and 11.2 mg gDCW−1 is presented, which is about 2.5 times higher than the next highest result (489 mg L−1) reported by Freivalds et al. [32].
We also demonstrate that the PI-algorithm-based residual methanol concentration control was able to maintain the selected set-point concentrations in the explored range; however, control quality was sensitive to the values of selected control parameters KP and τI.
The produced recombinant HBcAg molecules were successfully purified using a modified purification protocol, and the presence of highly immunogenic, correctly formed and homogenous HBcAg-VLPs with an estimated purity of 90% was confirmed using electron microscopy.
A relatively simple and robust bioreactor model to portray cell biomass (Xv, Xd and X), substrate (Gly. and MeOH), reactor volume (V), and product (HBcAg) dynamics during a P. pastoris GS115 cultivation at both limiting (0.1 g L−1) and residual (1.0 and 2.0 g L−1) methanol concentrations is presented. A rare attempt to model residual methanol concentration during the induction phase is also presented. Relatively easy implementation and parameter identification enables this approach to also be suitable for practical applications. The presented model could be further used also for model-based optimization and software-sensor development of the investigated process.
Abbreviations
- HBcAg:
-
Hepatitis B core Antigen
- DO:
-
Dissolved Oxygen
- OD:
-
Optical Density at 590 nm
- DCW:
-
Dry Cell Weight
- WCW:
-
Wet Cell Weight
- GMP:
-
Good Manufacturing Practice
- PID:
-
Proportional, Integral and Derivative control parameters
- PLC:
-
Programmable Logic Controller
- HPLC:
-
High-Performance Liquid Chromatography
- GC:
-
Gas Chromatography
- SCADA:
-
Supervisory Control and Data Acquisition interface
- Gly:
-
Glycerol
- MeOH:
-
Methanol
- Xv :
-
Vegetative cell biomass concentration [g L−1]
- Xd :
-
Dead (non-vegetative) cell biomass concentration [g L−1]
- X:
-
Total cell biomass concentration [g L−1]
- S:
-
Substrate concentration [g L−1]
- P:
-
Product concentration [g L−1]
- V:
-
Cultivation medium volume [L]
- τI :
-
Integral time constant [min.]
- Kp :
-
Proportional gain parameter [L2 g−1 h−1]
- K1, K2, k1, k2 :
-
Model parameters
- YATP :
-
ATP yield coefficient [g mol−1]
- YPX :
-
Specific product yield coefficient [g g−1]
- P/O:
-
Oxidative phosphorylation effectiveness coefficient
- φ:
-
Fraction of formaldehyde oxidised to formate
- mATP:
-
ATP maintenance coefficient [mol g−1 h−1]
- qG :
-
Specific glycolysis rate [mol g−1 h−1]
- qAc :
-
Specific acetyl-CoA production rate [mol g−1 h−1]
- qO2 :
-
Specific oxygen uptake rate [mol g−1 h−1]
- qGly :
-
Specific glycerol uptake rate [g g−1 h−1]
- qGly,R :
-
Specific glycerol uptake rate according to the Regulator model [g g−1 h−1]
- qGly,M :
-
Specific glycerol uptake rate according to the Monod model [g g−1 h−1]
- qmin,Gly :
-
Minimum specific glycerol uptake rate [g g−1 h−1]
- qmax,Gly :
-
Maximum specific glycerol uptake rate [g g−1 h−1]
- qMeOH :
-
Specific methanol uptake rate [g g−1 h−1]
- qMeOH,max :
-
Maximum specific methanol uptake rate [g g−1 h−1]
- qP :
-
Specific product formation rate [g g−1 h−1]
- SGly :
-
Glycerol concentration in cultivation medium [g L−1]
- SMeOH :
-
Methanol concentration in cultivation medium [g L−1]
- SF :
-
Substrate concentration in feed [g L−1]
- M:
-
Substrate molar mass [g mol−1]
- KGly :
-
Glycerol saturation constant [g L−1]
- KMeOH :
-
Methanol saturation constant [g L−1]
- Ki :
-
Methanol inhibition constant [g L−1]
- Kd :
-
Cell-specific death rate [h−1]
- μ :
-
Specific biomass growth rate [h−1]
- Fevp :
-
Evaporation rate [L h−1]
- Fs :
-
Substrate feed rate [L h−1]
- FNH3 :
-
Base addition rate [L h−1]
- FAF :
-
Antifoam addition rate [L h−1]
- Fsmpl :
-
Sampling rate [L h−1]
References
Roldão A, Mellado MCM, Castilho LR et al (2010) Virus-like particles in vaccine development. Expert Rev Vaccines 9:1149–1176. https://doi.org/10.1586/erv.10.115
Milich DR, McLachlan A, Moriarty A, Thornton GB (1987) Immune response to hepatitis B virus core antigen (HBcAg): localization of T cell recognition sites within HBcAg/HBeAg. J Immunol 139:1223–1231
Moradi Vahdat M, Hemmati F, Ghorbani A et al (2021) Hepatitis B core-based virus-like particles: A platform for vaccine development in plants. Biotechnol Reports 29:e00605. https://doi.org/10.1016/j.btre.2021.e00605
Pumpens P (2008) Construction of Novel Vaccines on the Basis of Virus-Like Particles. In: Khudyakov YE (ed) Medicinal Protein Engineering. CRC Press, Boca Raton, pp 205–248
Pumpens P, Grens E (2016) The true story and advantages of the famous hepatitis B virus core particles: Outlook 2016. Mol Biol 50:558–576. https://doi.org/10.7868/S0026898416040091
Nooraei S, Bahrulolum H, Hoseini ZS et al (2021) Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J Nanobiotechnology 19:1–27. https://doi.org/10.1186/s12951-021-00806-7
Schwartz L, Brown GV, Genton B, Moorthy VS (2012) A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar J 11:1–22. https://doi.org/10.1186/1475-2875-11-11
Laurens MB (2019) RTS, S/AS01 vaccine (MosquirixTM): an overview. Hum Vaccines Immunother 16:480–489. https://doi.org/10.1080/21645515.2019.1669415
Whelan MA, Field RA (2016) Vaccines based on Hepatitis B core antigens
Xunlong S (2016) Chimeric hepatitis B virus core antigen therapeutic vaccine of a kind of targeting and uses thereof
Wang L (2013) HLA-DR9 restrictive regulatory T cell epitope of Hepatitis B virus core antigen and e antigen and application thereof
Ma Y (2016) Application of HBcAg (hepatitis B core antigen) virus-like particle serving as cancer therapeutic vaccine carrier
Owczarek B, Gerszberg A, Hnatuszko-Konka K (2019) A Brief Reminder of Systems of Production and Chromatography-Based Recovery of Recombinant Protein Biopharmaceuticals. Biomed Res Int 2019:4216060. https://doi.org/10.1155/2019/4216060
Cregg JM (2000) Recombinant protein expression in Pichia pastoris. Appl Biochem Biotechnol - Part B Mol Biotechnol 16:23–52. https://doi.org/10.1385/MB:16:1:23
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270. https://doi.org/10.1002/yea.1208
Looser V, Bruhlmann B, Bumbak F et al (2015) Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnol Adv 33:1177–1193. https://doi.org/10.1016/j.biotechadv.2015.05.008
Liu WC, Inwood S, Gong T et al (2019) Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol 39:258–271. https://doi.org/10.1080/07388551.2018.1554620
Yang Z, Zhang Z (2018) Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: A review. Biotechnol Adv 36:182–195. https://doi.org/10.1016/j.biotechadv.2017.11.002
Potvin G, Ahmad A, Zhang Z (2012) Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: A review. Biochem Eng J 64:91–105. https://doi.org/10.1016/j.bej.2010.07.017
Gurramkonda C, Adnan A, Gäbel T et al (2009) Simple high-cell density fed-batch technique for high-level recombinant protein production with Pichia pastoris: Application to intracellular production of Hepatitis B surface antigen. Microb Cell Fact 8:1–8. https://doi.org/10.1186/1475-2859-8-13
Hong F, Meinander NQ, Jönsson LJ (2002) Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol Bioeng 79:438–449. https://doi.org/10.1002/bit.10297
Cos O, Ramon R, Montesinos JL, Valero F (2006) A simple model-based control forPichia pastoris allows a more efficient heterologous protein production bioprocess. Biotechnol Bioeng 95:145–154. https://doi.org/10.1002/bit.21005
Pla IA, Damasceno LM, Vannelli T et al (2006) Evaluation of Mut+ and MutS Pichia pastoris phenotypes for high level extracellular scFv expression under feedback control of the methanol concentration. Biotechnol Prog 22:881–888. https://doi.org/10.1021/bp060012+
Parliament European (2003) Commission Directive 2003/94/EC of 8 October 2003 laying down the principles and guidelines of good manufacturing practice in respect of medicinal products for human use and investigational medicinal products for human use
Food and Drug Administration (2004) Guidance for Industry, PAT-A Framework for Innovative Pharmaceutical Development, Manufacturing and Quality Assurance
Grigs O, Galvanauskas V, Dubencovs K, et al (2016) Model Predictive Feeding Rate Control in Conventional and Single-use Lab-scale Bioreactors: A Study on Practical Application. Chem Biochem Eng Q J 30:47–60. https://doi.org/10.15255/CABEQ.2015.2212
Jahic M, Rotticci-Mulder J, Martinelle M et al (2002) Modeling of growth and energy metabolism of Pichia pastoris producing a fusion protein. Bioprocess Biosyst Eng 24:385–393. https://doi.org/10.1007/s00449-001-0274-5
Ren HT, Yuan JQ, Bellgardt KH (2003) Macrokinetic model for methylotrophic Pichia pastoris based on stoichiometric balance. J Biotechnol 106:53–68. https://doi.org/10.1016/j.jbiotec.2003.08.003
Niu H, Daukandt M, Rodriguez C et al (2013) Dynamic modeling of methylotrophic Pichia pastoris culture with exhaust gas analysis: From cellular metabolism to process simulation. Chem Eng Sci 87:381–392. https://doi.org/10.1016/j.ces.2012.11.006
Barrigon JM, Valero F, Montesinos JL (2015) A macrokinetic model-based comparative meta-analysis of recombinant protein production by Pichia pastoris under AOX1 promoter. Biotechnol Bioeng 112:1132–1145. https://doi.org/10.1002/bit.25518
Zhang W, Bevins MA, Plantz BA et al (2000) ModelingPichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy-chain fragment C of botulinum neurotoxin, serotype A. Biotechnol Bioeng 70:1–8. https://doi.org/10.1002/1097-0290(20001005)70:1%3c1::AID-BIT1%3e3.0.CO;2-Y
Freivalds J, Dislers A, Ose V et al (2011) Highly efficient production of phosphorylated hepatitis B core particles in yeast Pichia pastoris. Protein Expr Purif 75:218–224. https://doi.org/10.1016/j.pep.2010.09.010
Invitrogen Corporation (2002) Pichia Fermentation Process Guidelines. https://tools.thermofisher.com/content/sfs/manuals/pichiaferm_prot.pdf
Grigs O, Bolmanis E, Galvanauskas V (2021) Application of In-Situ and Soft-Sensors for Estimation of Recombinant P. pastoris GS115 Biomass Concentration: A Case Analysis of HBcAg (Mut+) and HBsAg (MutS) Production Processes under Varying Conditions. Sensors 21:1268. https://doi.org/10.3390/s21041268
Ouchterlony O (1966) The antigenic pattern of immunoglobulins. G Mal Infett Parassit 18:942–948
Jackson JV, Edwards VH (1975) Kinetics of substrate inhibition of exponential yeast growth. Biotechnol Bioeng 17:943–964. https://doi.org/10.1002/bit.260170702
National Institutes of Health PubChem database. https://pubchem.ncbi.nlm.nih.gov/
Berza I, Dishlers A, Petrovskis I et al (2013) Plasmid dimerization increases the production of hepatitis B core particles in E. coli. Biotechnol Bioprocess Eng 18:850–857. https://doi.org/10.1007/s12257-013-0188-5
Peng M, Chen M, Ling N et al (2006) Novel vaccines for the treatment of chronic HBV infection based on mycobacterial heat shock protein 70. Vaccine 24:887–896. https://doi.org/10.1016/j.vaccine.2005.12.050
Jing XD, Wei H, Li H (2018) A kind of expression and purification method of HBcAg
Li ZX, Hong GQ, Hu B et al (2007) Suitability of yeast- and Escherichia coli-expressed hepatitis B virus core antigen derivatives for detection of anti-HBc antibodies in human sera. Protein Expr Purif 56:293–300. https://doi.org/10.1016/j.pep.2007.08.004
Watelet B, Quibriac M, Rolland D et al (2002) Characterization and diagnostic potential of hepatitis B virus nucleocapsid expressed in E. coli and P. pastoris. J Virol Methods 99:99–114. https://doi.org/10.1016/S0166-0934(01)00385-8
Rolland D, Gauthier M, Dugua JM et al (2001) Purification of recombinant HBc antigen expressed in Escherichia coli and Pichia pastoris: Comparison of size-exclusion chromatography and ultracentrifugation. J Chromatogr B Biomed Sci Appl 753:51–65. https://doi.org/10.1016/S0378-4347(00)00538-7
Zhang Y, Liu Y, Zhang B et al (2021) In vitro preparation of uniform and nucleic acid free hepatitis B core particles through an optimized disassembly-purification-reassembly process. Protein Expr Purif 178:105747. https://doi.org/10.1016/j.pep.2020.105747
Letourneur O, Watelet B (2001) HBcAg expression and diagnostic and therapeutic uses
Acknowledgements
The authors would like to acknowledge the contribution of Inara Akopjana for preparing seed inoculation cultures and performing SDS-PAGE analysis, Janis Bogans for performing chromatography runs, Dr. Juris Jansons for providing the electron microscopy images, and Rita Skerbaka for the off-line glycerol and methanol analysis. This research was funded by the European Regional Development Fund (ERDF) and Latvian State Project Agreement No. 1.1.1.2/16/I/001 research application (Grant) No. 1.1.1.2/VIAA/1/16/186.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bolmanis, E., Grigs, O., Kazaks, A. et al. High-level production of recombinant HBcAg virus-like particles in a mathematically modelled P. pastoris GS115 Mut+ bioreactor process under controlled residual methanol concentration. Bioprocess Biosyst Eng 45, 1447–1463 (2022). https://doi.org/10.1007/s00449-022-02754-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02754-4