Abstract
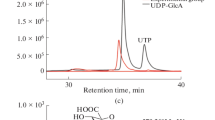
Five recombinant Escherichia coli extracts harboring overexpressed galactokinase, galactose-1-phosphate uridyltransferase, UDP-glucose pyrophophorylase, UMP kinase, and acetate kinase (AK) were utilized for the production of UDP-galactose (UDP-Gal). We analyzed the parameters which limit the yield of UDP-Gal in the reaction, and the reaction was optimized by increasing the concentration of AK. AK was used for the ATP regeneration as well as the conversion of UDP to UTP. The activities of four overexpressed enzymes were identically fixed, and then we increased the activity of AK to 20 times higher than others. The extracts catalyzed the production of UDP-Gal from UMP (10 mM), galactose (12 mM), ATP (1 mM), and acetyl phosphate (40 mM). As the result of the reaction, the conversion yield of UDP-Gal reached to 95% from 10 mM UMP.




Similar content being viewed by others
Abbreviations
- Acetyl Pi:
-
Acetyl phosphate
- ADP:
-
Adenosine 5′-diphosphate
- ATP:
-
Adenosine 5′-triphosphate
- CMP-NeuAc:
-
Cytidine 5′-monophosphate
- dNTP:
-
Deoxynucleoside-triphosphate
- E. coli :
-
Escherichia coli
- Gal:
-
Galactose
- Glc:
-
Glucose
- Pi:
-
Phosphate
- PPi:
-
Pyrophosphate
- UDP:
-
Uridine 5′-diphosphate
- UDP-Gal:
-
Uridine 5′-diposphogalactose
- UDP-Glc:
-
Uridine 5′-diphosphoglucose
- UMP:
-
Uridine 5′-monophosphate
- UTP:
-
Uridine 5′-triphosphate
References
Cummings RD, Merkle RK, Stults NL (1989) Separation and analysis of glycoprotein oligosaccharides. Methods Cell Biol 32:141–183
Rudd PM, Leatherbarrow RJ, Rademacher TW, Dwek RA (1991) Diversification of the IgG molecule by oligosaccharides. Mol Immunol 28:1369–1378
Spiro RG (1973) Glycoproteins. Adv Protein Chem 27:349–467
Montreuil J (1980) Primary structure of glycoprotein glycans: basis for the molecular biology of glycoproteins. Adv Carbohydr Chem Biochem 37:157–223
Liu Z, Zhang J, Chen X, Wang PG (2002) Combined biosynthetic pathway for de novo production of UDP-galactose: catalysis with multiple enzymes immobilized on agarose beads. Chembiochem 3:348–355
Oh JM, Lee SG, Kim BG, Sohng JK, Liou K, Lee HC (2003) One-pot enzymatic production of dTDP-4-keto-6-deoxy-d-glucose from dTMP and glucose-1-phosphate. Biotechnol Bioeng 84:452–458
Liu JL, Shen GJ, Ichikawa Y, Rutan JF, Zapata G, Vann WF, Wong CH (1992) Overproduction of CMP–sialic acid synthetase for organic synthesis. J Am Chem Soc 114:3901–3910
Lee JO, Yi JK, Lee SG, Takahashi S, Kim BG (2004) Production of N-acetylneuraminic acid from N-acetylglucosamine and pyruvate using recombinant human renin binding protein and sialic acid aldolase in one pot. Enzyme Microb Technol 35:121–125
Lee SG, Lee JO, Yi JK, Kim BG (2002) Production of cytidine 5′-monophosphate N-acetylneuraminic acid using recombinant Escherichia coli as a biocatalyst. Biotechnol Bioeng 80:516–524
Koizumi S, Endo T, Tabata K, Ozaki A (1998) Large-scale production of UDP-galactose and globotriose by coupling metabolically engineered bacteria. Nat Biotechnol 16:847–850
Holden HM, Rayment I, Thoden JB (2003) Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 278:43885–43888
Hwang S, Choi CY, Lee EY (2008) One-pot biotransformation of racemic styrene oxide into (R)-1,2-phenylethandiol by two recombinant microbial epoxide hydrolases. Biotechnol Bioproc Eng 13:453–457
Ryll T, Wagner R (1991) Improved ion-pair high-performance liquid chromatographic method for the quantification of a wide variety of nucleotides and sugar-nucleotides in animal cells. J Chromatogr 570:77–88
Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC (2001) Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem 293:129–137
Doyle JD, Parsons SA (2002) Struvite formation, control and recovery. Water Res 36:3925–3940
Stratful I, Scrimshaw MD, Lester JN (2001) Conditions influencing the precipitation of magnesium ammonium phosphate. Water Res 35:4191–4199
Acknowledgments
This research was supported by a grant (M10417060004-04N1706-00410) from Korea Biotech R&D Group of Next-generation growth engine project of the Ministry of Education, Science and Technology, Republic of Korea and the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MOST) (No. R0A-2007-000-10007-0).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, JH., Chung, SW., Lee, HJ. et al. Optimization of the enzymatic one pot reaction for the synthesis of uridine 5′-diphosphogalactose. Bioprocess Biosyst Eng 33, 71–78 (2010). https://doi.org/10.1007/s00449-009-0365-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-009-0365-2