Abstract
Spatial capture–recapture modelling (SCR) is a powerful tool for estimating density, population size, and space use of elusive animals. Here, we applied SCR modelling to non-invasive genetic sampling (NGS) data to estimate red fox (Vulpes vulpes) densities in two areas of boreal forest in central (2016–2018) and southern Norway (2017–2018). Estimated densities were overall lower in the central study area (mean = 0.04 foxes per km2 in 2016, 0.10 in 2017, and 0.06 in 2018) compared to the southern study area (0.16 in 2017 and 0.09 in 2018). We found a positive effect of forest cover on density in the central, but not the southern study area. The absence of an effect in the southern area may reflect a paucity of evidence caused by low variation in forest cover. Estimated mean home-range size in the central study area was 45 km2 [95%CI 34–60] for females and 88 km2 [69–113] for males. Mean home-range sizes were smaller in the southern study area (26 km2 [16–42] for females and 56 km2 [35–91] for males). In both study areas, detection probability was session-dependent and affected by sampling effort. This study highlights how SCR modelling in combination with NGS can be used to efficiently monitor red fox populations, and simultaneously incorporate ecological factors and estimate their effects on population density and space use.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reliable information on animal population status, including population size and density, is crucial for wildlife research and management (Kämmerle et al. 2018). However, estimating population size and density is challenging. This is especially true for predators, because they often occur at low densities, are elusive, and inhabit areas that may also be difficult to survey due to inaccessibility or rough terrain (Kery et al. 2011). Predators are also often of management concern due to their conservation status or conflict potential with humans through direct threat, depredation of livestock, competition for game species (Estes 1996), or spreading pathogens (Moore et al. 2010b).
The red fox (Vulpes vulpes) is a highly adaptable and opportunistic mesopredator with a broad ecological niche and variable diet, including both wild and domestic vertebrates (Dell'Arte et al. 2007; Killengreen et al. 2011). It is the most widely distributed carnivore in the world and is commonly found in a wide array of habitats. It is also considered invasive or overabundant across much of its geographic range (Larivière and Pasitschniak‐Arts 1996). The species’ ongoing geographic expansion is of management concern due to deleterious effects on populations of other species. This includes intraguild competition with arctic fox (Vulpes lagopus; Frafjord et al. 1989), and predation on threatened species like the lesser white-fronted goose (Anser erythropus; Aarvak et al. 2017), and game species like forest birds (Doherty et al. 2016; Jahren 2017; Skrede 2016; Smedshaug et al. 1999). The red fox is also a vector of zoonotic pathogens that can pose risks for domestic animals and humans (Hodžić et al. 2016; Víchová et al. 2018; Laurimaa et al. 2016). Despite the importance of the red fox for wildlife management, a few practical methods are available for estimating population size and densities, required to evaluate effects of management actions (Wegge et al. 2019). Because direct observation of the red fox is difficult (Vine et al. 2009), methods used to monitor red fox populations have mainly been based on indirect measures, including culling indices (Smedshaug et al. 1999), snow tracking (Wegge and Rolstad 2011), fecal counts (Cavallini 1994; Webbon et al. 2004), mapping of active dens (Lindström 1989; Lindström et al. 1994), and camera trap visits (Hamel et al. 2013; Henden et al. 2014). These methods assume that the measured indices are directly proportional to the population parameter of interest, be it population size or density. This relationship is, however, often unknown, and thus, the reliability of these methods is difficult to evaluate (O'Connell et al. 2010; Sollmann et al. 2013).
An alternative approach is capture–recapture (CR) methodology. CR methods are widely used for estimating animal population parameters (Silvy 2012). CR uses multiple captures of the same individual, identified by natural or artificial means, to make extended inferences at the population level. An important advantage of these methods is their ability to account for imperfect and variable detection probability (Amstrup et al. 2010; Royle and Young 2008). Conventional CR, however, exhibits difficulty associated with estimating population density due to movements of animals into and out of the study area, which often leads to erroneous inferences (Royle and Young 2008; Royle et al. 2018).
Unlike conventional CR, spatial capture–recapture (SCR) incorporates a spatially explicit component in the model that accounts for spatial heterogeneity in detection probability of individuals. SCR can therefore estimate density as an explicit parameter (Royle et al. 2013). In addition, SCR models allow for the incorporation of ecological factors, such as sex or habitat characteristics, and estimate effects of these on population density and animal space use. SCR is also well suited for use in combination with non-invasive sampling methods, such as camera trapping and non-invasive genetic sampling (NGS) data (Mumma et al. 2015; Royle et al. 2013). NGS in combination with SCR methods has recently become a popular tool to monitor wide-ranging carnivores at large scales (Bischof et al. 2020). Recent studies also support use of these methods to monitor mesopredators when applied at appropriate spatial scales (Morin et al. 2016; Wegge et al. 2019).
The goal of the present study is to assess the combination of non-invasive genetic sampling with spatial capture–recapture for estimating red fox density, and explore the role of individual and spatial variables on density, space use, and detectability. We use data from two different study areas in Norway with different habitat and climate characteristics.
Materials and methods
Study areas
The first study area (“Lierne”) was established in Lierne, Trøndelag in central Norway (64.353° N, 13.659° E; Fig. 1A), where a pilot study was conducted in 2016. It consists of an undulating terrain between 500 and 950 m a.s.l. with mixed forests and protruding unforested crests, and a mean forest cover of 50%. Norway spruce (Picea abies) dominates the forests with interspersed Birch (Betula spp.) and Scots pine (Pinus sylvestris) (Moen 1998). Parts of the study area are subjected to commercial clear-cut forestry, and small settlements are scattered along the main road going through the study area. Parts of the region are used by semi-domestic reindeer (Rangiferus tarandus) for perennial pastures in addition to moose (Alces alces) and roe deer (Capreolus capreolus), and a diverse carnivore community, including arctic fox (Vulpes lagopus), wolverine (Gulo gulo), brown bear (Ursus actos), lynx (Lynx lynx), and pine marten (Martes martes; Gomo et al. 2017, 2020).
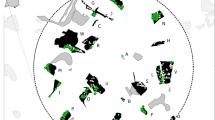
Map of the two 225 km2 study areas in A Lierne in central Norway and B Skrim in southern Norway. The study areas are shown with a 5 × 5 km grid with locations of all fecal, urine, and hair samples included in genetic analysis to identify individual red foxes, and subsequently used for estimating red fox density. Samples of the same colour represent samples from the same individual. Inset panels show each study area’s location in Norway
The second study area (“Skrim”) was established in 2017 near Skrim, Viken in southern Norway (59.391° N, 9.590° E; Fig. 1B). This study area is located between 400 and 675 m a.s.l., and is comparable to Lierne in terms of species composition (Østbye et al. 1989) and forestry practice (Moen 1998), but with denser forest cover (85%), rougher topography, and no unforested crests. The fauna in Skrim is less documented but comparable to Lierne, though the study area is located outside the range of wolverine, arctic fox, and reindeer. Human occupancy along the main roads is similar in both study areas (Norwegian Mapping Authority 2020), but the human population in adjacent settlements is substantially higher in Skrim municipality which includes a city (Skien) of 55 000 inhabitants. By contrast, the population of the entire municipality of Lierne is 1355 inhabitants (Statistics Norway 2020). Both study areas are 15 × 15 km (225 km2; Fig. 1).
Data collection
Scats, urine, and hair from red fox were collected during February and March in 2016, 2017 and 2018 in Lierne, and in 2017 and 2018 in Skrim. Both study areas were divided into 5 × 5 km grids to guide the allocation of search effort. Sampling was predominantly done by the same local hunters each year and primarily focused along snow covered dirt roads, snowmobile tracks, and skiing tracks. Urine samples were collected by placing spruce sticks (40–60 cm in length) for foxes to urinate on at an interval of approximately 500 m along sampled roads and tracks. Each transect was sampled at least twice each year. Scat, urine, and hair samples were handled with gloves and plastic cutlery to avoid contamination of DNA, and placed in plastic vials containing silica gel or urine preservative fluid and paper envelopes, respectively, for preservation of DNA and storage for later analysis. All samples were dated and corresponding UTM coordinates were recorded with a handheld GPS unit.
DNA extraction, amplification, and genotyping
The genetic analyses were undertaken at the Norwegian Institute for Nature Research (NINA) in Trondheim, Norway. DNA was extracted from 314 scat, 448 urine, and 23 hair samples (Table S1) using the FastDNA™ Spin Kit for Soil, the Norgen Biotek Urine DNA Isolation Kit (Slurry Format) and the Maxwell® 16 Tissue DNA Purification Kit, respectively, following the manufacturer’s protocols. To confirm red fox samples, two PCR runs followed by capillary electrophoresis were performed for each sample using the species identification method described by Dalén et al. (2004). Samples from other species than red fox were excluded from further analysis. All confirmed red fox samples were genotyped with 14 microsatellite markers, including a marker for sex determination (Moore et al. 2010a). To account for genotyping errors in low-quality samples (Fig. S1 and S2), three replicates per sample and marker were applied.
Consensus genotypes were assigned to each sample based on consistency across all three replicates for homozygote markers and at least two for heterozygotes. This procedure minimizes the risk of genotyping errors caused by allelic dropout and false alleles (Taberlet et al. 1996). To identify reliable genotypes, we assigned each sample a quality index (QI), calculated as the proportion of consistent gene scores across all three replicates (Miquel et al. 2006). Samples with a mean QI of 0.70 or above were retained for subsequent individual identification. Finally, we assigned identities using Allelematch, an R package for identifying unique multilocus genotypes where genotyping error and missing data may be present (Galpern et al. 2012), in R version 3.6.0 (R core team 2019).
Spatial capture–recapture
General description
We estimated red fox densities for each study area using spatial capture–recapture (SCR) models. SCR models are hierarchical models composed of a submodel for the distribution of individuals in space, i.e., density (D), and a submodel for the detection of these same individuals, conditional on their location. SCR models assume that animals move around a central point referred to as the activity centre (AC). Density is modelled as the distribution of ACs over an area referred to as the state space that encompasses the surveyed area surrounded by a buffer large enough to include the AC location of any individual that could have been exposed to sampling (Royle et al. 2013). Density may be modelled as a function of spatially explicit covariates (Borchers and Efford 2008). SCR models usually assume that the detection probability of an individual declines with distance to an individual's AC. The most common detection model is the half-normal function, which has two parameters. The scale parameter (σ) describes how fast the detection probability decreases with distance, and the baseline detection probability (p0) describes the probability to detect an individual at the exact location of its AC. Both the scale parameter and the baseline detection probability can be related to different individual or spatial covariates to account for potential heterogeneity in detection (Royle et al. 2013). The detection model also implies a model of space use that is closely linked with home-range size through 1) movement of an individual about its home-range and 2) detection being proportional to space use in the vicinity of a detector. We can thus use SCR models to derive sex-specific home-range size estimates (i.e., the circular area encompassed by the 95% vertex of the utilization distribution) directly from the scale parameter σ using the Chi-square distribution with two degrees of freedom (Royle et al. 2013).
State space, detectors, and SCR data
Models were run separately for each study area, and therefore, the state space and potential detection locations, i.e., detectors, were also study area-specific. Detectors were defined as the centres of 500 × 500 m grid cells covering each 225 km2 study area (N = 900; Fig. S3 and S4). The state space for each study area was defined as a grid of 500 × 500 m cells covering the area searched for DNA samples surrounded by an 8000 m buffer. The buffer width was calculated by multiplying by 4 the largest estimated σ in a preliminary analysis (Efford 2004).
Only samples found within the spatial bounds of the study areas (Fig. 1) for which coordinates, species, sex, and individual ID were available were considered a detection and assigned to the nearest detector. SCR datasets for each year and study area were built from the number of individual detections at each detector.
Model implementation and selection
We ran all models as multi-session spatial capture–recapture models using the oSCR package version 0.42.0 (Sutherland et al. 2019) in R version 3.6.0 (R core team 2019). The multi-session implementation allowed us to use data from different sessions, in this case years, in a single statistical model. This increases reliability and estimates effects on different parameters either jointly across sessions or independently (Sutherland et al. 2019).
We first constructed a simple multi-session model based on inherent study design specifications. We included session-specific density and detection probability to obtain year-specific estimates. To control for variable search effort along search transects, we also included a detector-specific covariate on p0, defined as the total length of registered GPS search tracks within each detector grid cell. To test for the effect of multiple covariates on red fox density, detection, and space use, we built and compared 16 extensions of the simple model, based on all possible combinations of the covariates of interest explained below.
To test for a relationship between red fox density and available forest habitat, as suggested by previously reported habitat preferences of the red fox (Cagnacci et al. 2004; Svendsen 2016; Van Etten et al. 2007), we considered an effect of forest cover as a spatial predictor of density (Molina et al 2017). Proportion of forest cover for each state-space grid cell was extracted in QGIS version 3.10 (QGIS Development Team 2019) based on maps at scales between 1:25 000 and 1:100 000 (Norwegian Mapping Authority 2020).
As density in SCR translates to the distribution of individual home ranges across the landscape, i.e., second-order habitat selection (Everatt et al. 2015), proportion of forest cover was defined as the average forest cover in a 1000 m radius around each raster cell.
To test for sexual dimorphism in red fox space use, we included sex as a predictor of σ and p0. Some studies report home-range size of the red fox to differ between sexes (Drygala and Zoller 2013), while other studies report no significant sex differences (Svendsen 2016; Walton et al. 2017). Because parameterization of the scale parameter affects baseline detection probability and vice versa (Efford and Mowat 2014), the sex effect was always tested simultaneously on σ and p0.
Furthermore, to account for additional sources of variation in detectability, we included road length and forest cover as predictors of p0. The inclusion of the road covariate was motivated by evidence suggesting that mesopredators often travel along roads in winter to conserve energy (Crête and Larivière 2003), while forest cover was included to test whether detectability was higher in open vs. covered areas. As an individual’s detection probability is tightly linked with space use within its home range, i.e., fourth-order habitat selection, all spatial covariates on p0 were extracted at the detector grid scale. Road length was thus defined as the total length of roads and forest cover as the average forest cover within each detector grid cell, based on maps at scales between 1:25 000 and 1:100 000 (Norwegian Mapping Authority 2020) Table 1.
Fitted models were subjected to post-processing and model selection using functionality provided in the oSCR package. Model selection was performed using the Akaike Information Criterion (AIC; Burnham and Anderson 2002). For each study area, only the model with the lowest AIC value was retained for estimating density, population size, home range, and covariate effects (Table 2 and 3). Predicted values from the retained fitted models, including red fox density, realized density maps, and population size, were produced using functionality provided in the oSCR package (Sutherland et al. 2019). Additional detail is provided in Online Resource 2.
Results
NGS samples
Out of 502 total samples collected in Lierne, 383 were confirmed as red fox, of which 275 samples were successfully assigned reliable genotypes and individual IDs (for a breakdown by sample type, see Table S1). Successfully genotyped samples originated from 98 different individuals. The mean number of samples per individual was 2.23 (range: 1–8) in 2016, 2.57 (range: 1–12) in 2017, and 4.57 (range: 1–17) in 2018 (Table 1). Out of 283 total samples collected in Skrim, 223 were confirmed as red fox, of which 103 were successfully assigned reliable genotypes and individual IDs. Successfully genotyped samples originated from 39 different individuals. The mean number of samples per individual was 1.72 (range: 1–4) in 2017 and 2.40 (range: 1–6) in 2018 (Table 1). A summary of the samples included in the SCR analysis is provided in Table S4.
Model selection
The top model for Lierne included forest cover and year effects on density, year and search effort effects on baseline detection probability, and sex effects on both baseline detection probability and the scale parameter. For Skrim, the top model did not include an effect of forest cover on density. Baseline detection probability was influenced by year, search effort, and road length and fox sex influenced both baseline detection probability and the scale parameter.
Estimated population size and density
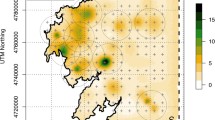
Estimated average red fox densities across the study area in Lierne (Fig. 1A) were 0.04 foxes per km2 in 2016, 0.10 in 2017, and 0.06 in 2018. Furthermore, density was predicted to increase with forest cover (βforest = 2.91 [95%CI 0.14–5.67]; Fig. 2A; Table S5). Mean-estimated population size within the original 225 km2 study area in Lierne (Fig. 1A) was 9 foxes in 2016, 22 in 2017, and 14 in 2018.
Estimated red fox density from the best-supported spatial capture–recapture models based on non-invasive DNA samples from Lierne, central Norway (2016–2018) and Skrim, southern Norway (2017–2018). The best-supported model for Lierne included effects of year and forest cover on red fox density (A) and only differences between years in Skrim (B). Plots present the mean predicted densities (colored lines in A and point in B) and associated 95% confidence intervals (dashed lines in A and whiskers in B)
Estimated average red fox densities across the study area in Skrim (Fig. 1B) were 0.16 and 0.09 foxes per km2 in 2017 and 2018, respectively (Fig. 2B). These estimates corresponded to population sizes of 36 foxes and 20 foxes within the 225 km2 study area in 2017 and 2018, respectively (Fig. 1B). Mean-estimated densities for each study area per year are shown in Figs. 3, 4, and 5.
Estimated red fox baseline detection probability (p0) from the best-supported spatial capture–recapture models based on non-invasive DNA samples from Lierne, central Norway (2016–2018) and Skrim, southern Norway (2017–2018). The best-supported model for Lierne included effects of year and search effort on red fox baseline detection probability (A), while the best model for Skrim included effects of year, search effort (B), and road length (C). Both models also included a sex effect; presented are predicted p0 values for males. Coloured lines represent the mean predicted values and dashed lines represent the associated 95% confidence intervals
Estimated red fox scale parameter (σ) from the best-supported spatial capture–recapture models based on non-invasive DNA samples from Lierne (coloured circles) and Skrim (coloured squares), Norway. The best-supported model for both Lierne and Skrim included a difference between sexes. Dots represent the mean values and whiskers represent the associated 95% confidence intervals
Detection and space use
In Lierne, baseline detection probability increased with search effort (βsearch = 0.76 [0.60–0.92]; Fig. 3A) and was higher for males than for females, albeit not significantly (βsexMale = 0.32 [ – 0.23–0.88]; Table S5). In Skrim, baseline detection probability increased with both search effort (βsearch = 0.33 [0.12–0.54]; Fig. 3B) and road length (βroad = 0.2 [0.03–0.43]; Fig. 3C). In addition, baseline detection probability was non-significantly higher for females than for males in Skrim (βsexMale = – 0.76 [ – 1.71–0.19]; Table S6).
In Lierne, σ estimates were 1.63 [1.38 -1.92] km for females and 2.13 [1.87–2.44] km for males (Fig. 4), which corresponded to home-range sizes of 50 [36–69] and 86 [66–112] km2 for females and males, respectively. In Skrim, σ estimates were 1.17 [0.91–1.49] km for females and 1.73 [1.36–2.19] km for males (Fig. 4), corresponding to home-range sizes of 26 [16–42] km2 and 56 [35–91] km2, respectively.
Discussion
Ecologists and wildlife managers have a keen interest in quantifying wildlife abundance across landscapes and identifying the drivers of spatial variation therein (Bischof et al. 2020). The ability to accomplish these goals has gained spatial capture–recapture analysis substantial popularity over the past decade (Royle et al. 2018). SCR has proven a particularly powerful tool when used in concert with non-invasive methods as this has allowed population estimation at a scale and with a level of detail that had until recently been unattainable (Bischof et al. 2020).
Despite being the most widespread carnivore species globally, there is a paucity of detailed information about red fox population densities and their determinants (Wegge et al. 2019). Using non-invasive genetic sampling and SCR analysis, we mapped the density of red foxes in two boreal forest landscapes in Norway over 3 years. Our study revealed that a combination of spatial and individual factors influences density, space use, and detection probability.
We estimated higher red fox densities in the southern study area (Skrim) compared to the forest in central Norway (Lierne). Higher altitude, higher latitude, a more continental climate, and lower winter temperatures make Lierne less productive than Skrim (Moen 1998). The difference in estimated red fox densities between the study areas may partially be attributed to difference in vegetation and climate (Walton et al. 2017), and winter severity as limiting factors on density (Bartoń and Zalewski 2007). Human land use and anthropogenic subsidies have also been suggested to be important drivers of red fox density (Gomo et al. 2017; Rød-Eriksen et al. 2020). Forest landscapes with high human settlement density are associated with higher red fox abundances, potentially driven by increased food availability of anthropogenic origin, and thus increased scavenging opportunities (Jahren et al. 2020; Rød-Eriksen et al. 2020). Due to a larger human population as well as more clusters of cabins in adjacent areas in Skrim compared to Lierne, differences in density estimates may partially reflect differences in human influence.
The two study areas also differed in terms of variables that predicted red fox density. We detected a significant positive effect of forest cover on red fox density in Lierne but not in Skrim. Boreal forests are important habitats for several prey species of red fox, including voles, shrews, and forest birds (Needham et al. 2014). Forests likely also provide important refuges in winter in contrast to more exposed alpine areas. The lack of an effect of forest cover in Skrim may reflect a paucity of evidence, perhaps because variation in forest cover was very low, with less open unforested areas like bogs and impediment (Fig. S4). We note that several candidate models were close in support based on AIC in both study areas (Tables 2 and 3). However, additional covariate effects included in these models did not explain enough variation to justify their inclusion and were therefore not interpreted as having an ecological effect (Arnold 2010).
SCR models allowed us to derive sex-specific home-range sizes. The approach implemented here relies on the assumption of a normally distributed circular home range, and we note that SCR-derived home-range estimates have been shown to be sensitive to misspecification of the detection function (Royle et al. 2014). Nevertheless, our home-range size estimates for both study areas were comparable to estimates from two recent GPS telemetry studies of red fox in similar habitat in Scandinavia. Svendsen (2016) reported mean red fox home-range size of 61 km2 [95% CI 25–105] for the region of Østerdalen, Innlandet, and Walton et al. (2017) reported mean home-range sizes of 52 km2 [95% CI 32–72] for the regions of Kolmården, Grimsö, and Hedemora in Sweden, and Hedmark in Norway. However, the variation in reported individual home-range estimates was significant in both studies. Walton et al. also reported home ranges up to four times larger in less-productive and high elevation landscapes compared to more productive and low elevation landscapes (2017). We found a similar pattern with smaller home ranges in the more productive lower elevation southern boreal forest (26 [16–42] km2 for females and 56 [35–91] km2 for males in Skrim), compared to Lierne’s less-productive higher elevation northern boreal forest (45 [34–60] km2 for females and 88 [69–113] km2 for males). In contrast to studies by Svendsen (2016) and Walton et al. (2017), which reported no differences in home range between males and females, our study found home-range estimates of males to be approximately twice the size of females in both study areas. This may reflect variation in space use related to breeding status of females, as reproductive females have been reported to have smaller home ranges (Henry et al. 2005). Some females may have started retreating to natal dens towards the end of the sampling period (Walton and Mattisson 2021), which would affect their home-range sizes. Furthermore, DNA sampling was partly done during the mating period (January–March), when male foxes likely roam around to cover several female home ranges (Cavallini 1996), which could contribute to the observed difference between males and females.
One important advantage of SCR is that it accounts for imperfect and variable detection of individuals. Though many count-based wildlife surveys assume complete detection of all individuals in a population, this assumption is almost always violated (Kellner and Swihart 2014). When not accounted for, imperfect detection can lead to erroneous inferences about density and its drivers (Gu and Swihart 2004). In addition, in most monitoring set-ups, detection probability differs amongst individuals in the population as a result of different exposure to detectors in relation to individual home-range locations (Efford and Mowat 2014). SCR models use this inherent heterogeneity in detectability to estimate individual activity centres and space-use patterns (Royle et al. 2013). In our study, variation in detection probability was also influenced by spatial predictors, including a positive effect of search effort effect in both areas, and a positive trend of an effect of road length in Skrim only (Table S6). Lack of an effect of roads in Lierne may be due to insufficient evidence, as roads were fewer and covered less of the study area (Fig. S3). Detection probability also differed between years. Given that the detection of individual animals depended on the genetic analysis of NGS samples, this may reflect variation in genotyping success rates between years (Table 1).
Forty-eight percent of all samples collected in our study contained DNA of sufficient quality for individual identification. The proportion of successfully genotyped samples was noticeably higher in Lierne compared to Skrim, and fecal samples had the highest genotyping success rates in both areas (Table 1 and S1, Fig. S1). Many factors, including sample age, temperature, moisture, and UV radiation, contribute to degradation and preservation of DNA (Hausknecht et al. 2007; Panasci et al. 2011; Piggott 2005; Woodruff et al. 2015). The reported differences in genotyping success rates may thus reflect differences in environmental conditions. Considering that the samples collected were of varying type and quality, the genotyping success rates reported here validate the NGS methods as viable for identifying individual foxes. However, we also want to highlight the possibilities of implementing other types of data into the SCR framework for future studies. Multiple data sources, such as recoveries of dead animals, can also be integrated in the SCR framework to increase the precision of estimates (Dupont et al. 2021). Several methods were recently proposed for incorporating detections of unidentified individuals, leading to more precise estimation (Jiménez et al. 2019, 2021; Tourani et al. 2020).
A similar study by Wegge et al. (2019) produced red fox density estimates using SCR, but argued that a main shortcoming of their study was a smaller sampled area (50 km2). Because the scale parameter relates to home-range size, parameter estimates including density are more likely to be biased when the sampled area is small relative to the range of individual movements in the study population, particularly if sample size is low (Sollmann et al. 2012; Sun et al. 2014). We thus want to reemphasize the importance of considering spatial detector configuration relative to the known space use and home-range size of the study species.
Though we obtained density estimates that varied between years, the closed population SCR approach applied here is less suitable for studying the drivers of inter-annual differences in red fox density. Recent developments of SCR methods include open population analyses that allow for studying red fox population dynamics over time, including estimating mortality and recruitment rates, as well as immigration and emigration (Morin et al. 2016). This would also make better use of the available data, as information on individual states is propagated between years (Ergon and Gardner 2014; Gardner et al. 2018; Milleret et al. 2020).
The combination of SCR and NGS methods provides a solid framework not only for estimating red fox density, but also to identify drivers thereof (e.g., productivity, snow depth, forest cover, and influence of human activity). If applied at larger scales in different habitats, e.g., mosaics of forest and farmland or arctic and alpine areas, this approach has the potential to provide new insight into the relative importance of various drivers of red fox population dynamics.
References
Aarvak T, Øien JI, Karvonen R (2017) Development and key drivers of the Fennoscandian Lesser White-fronted Goose population monitored in Finnish Lapland and Finnmark. Safeguarding the Lesser White-fronted Goose Fennoscandian population at key staging and wintering sites within the European flyway: 29–36
Amstrup SC, McDonald TL, Manly BF (eds) (2010) Handbook of capture–recapture analysis. Princeton University Press, Princeton
Arnold TW (2010) Uninformative parameters and model selection using Akaike’s Information Criterion. J Wildl Manag 74(6):1175–1178
Bartoń KA, Zalewski A (2007) Winter severity limits red fox populations in Eurasia. Glob Ecol Biogeogr 16(3):281–289
Bischof R, Milleret C, Dupont P, Chipperfield J, Tourani M, Ordiz A, de Valpine P, Turek D, Royle JA, Gimenez O, Flagstad Ø, Åkesson M, Svensson L, Brøseth H, Kindberg J (2020) Estimating and forecasting spatial population dynamics of apex predators using transnational genetic monitoring. Proc Natl Acad Sci 117(48):30531–30538
Borchers DL, Efford MG (2008) Spatially explicit maximum likelihood methods for capture–recapture studies. Biometrics 64(2):377–385
Burnham KP, Anderson DR (2002) A practical information-theoretic approach. Model selection and multimodel inference, 2nd edn. Springer, New York, pp 70–71
Cagnacci F, Meriggi A, Lovari S (2004) Habitat selection by the red fox Vulpes vulpes (L. 1758) in an Alpine area. Ethol Ecol Evol 16(2):103–116
Cavallini P (1994) Faeces count as an index of fox abundance. Acta Theriol 39:417
Cavallini P (1996) Ranging behaviour of red foxes during the mating and breeding seasons. Ethol Ecol Evol 8(1):57–65
Crête M, Larivière S (2003) Estimating the costs of locomotion in snow for coyotes. Can J Zool 81(11):1808–1814
Dalén L, Elmhagen B, Angerbjörn A (2004) DNA analysis on fox faeces and competition induced niche shifts. Mol Ecol 13(8):2389–2392
Dell’Arte GL, Laaksonen T, Norrdahl K, Korpimäki E (2007) Variation in the diet composition of a generalist predator, the red fox, in relation to season and density of main prey. Acta Oecologica 31(3):276–281
Doherty TS, Glen AS, Nimmo DG, Ritchie EG, Dickman CR (2016) Invasive predators and global biodiversity loss. Proc Natl Acad Sci 113(40):11261–11265
Drygala F, Zoller H (2013) Spatial use and interaction of the invasive raccoon dog and the native red fox in Central Europe: competition or coexistence? Eur J Wildl Res 59(5):683–691
Dupont P, Milleret C, Tourani M, Brøseth H, Bischof R (2021) Integrating dead recoveries in open-population spatial capture–recapture models. Ecosphere 12(7):e03571
Efford M (2004) Density estimation in live-trapping studies. Oikos 106(3):598–610
Efford MG, Mowat G (2014) Compensatory heterogeneity in spatially explicit capture–recapture data. Ecology 95(5):1341–1348
Ergon T, Gardner B (2014) Separating mortality and emigration: modelling space use, dispersal and survival with robust-design spatial capture–recapture data. Methods Ecol Evol 5(12):1327–1336
Estes JA (1996) Predators and ecosystem management. Wildl Soc Bull 24(3):390–396
Everatt KT, Andresen L, Somers MJ (2015) The influence of prey, pastoralism and poaching on the hierarchical use of habitat by an apex predator. African J Wildlife Res 45(2):187–196
Frafjord K, Becker D, Angerbjörn A (1989) Interactions between arctic and red foxes in Scandinavia—predation and aggression. Arctic 42(4):354–356
Galpern P, Manseau M, Hettinga P, Smith K, Wilson P (2012) Allelematch: an R package for identifying unique multilocus genotypes where genotyping error and missing data may be present. Mol Ecol Resour 12(4):771–778
Gardner B, Sollmann R, Kumar NS, Jathanna D, Karanth KU (2018) State space and movement specification in open population spatial capture–recapture models. Ecol Evol 8(20):10336–10344
Gomo G, Mattisson J, Hagen BR, Moa PF, Willebrand T (2017) Scavenging on a pulsed resource: quality matters for corvids but density for mammals. BMC Ecol 17(1):1–9
Gomo G, Rød-Eriksen L, Andreassen HP, Mattisson J, Odden M, Devineau O, Eide NE (2020) Scavenger community structure along an environmental gradient from boreal forest to alpine tundra in Scandinavia. Ecol Evol 10(23):12860–12869
Gu W, Swihart RK (2004) Absent or undetected? Effects of non-detection of species occurrence on wildlife–habitat models. Biol Cons 116(2):195–203
Hamel S, Killengreen ST, Henden JA, Yoccoz NG, Ims RA (2013) Disentangling the importance of interspecific competition, food availability, and habitat in species occupancy: recolonization of the endangered Fennoscandian arctic fox. Biol Cons 160:114–120
Hausknecht R, Gula R, Pirga B, Kuehn R (2007) Urine—a source for noninvasive genetic monitoring in wildlife. Mol Ecol Notes 7(2):208–212
Henden JA, Stien A, Bårdsen BJ, Yoccoz NG, Ims RA (2014) Community-wide mesocarnivore response to partial ungulate migration. J Appl Ecol 51(6):1525–1533
Henry C, Poulle ML, Roeder JJ (2005) Effect of sex and female reproductive status on seasonal home range size and stability in rural red foxes (Vulpes vulpes). Ecoscience 12(2):202–209
Hodžić A, Alić A, Klebić I, Kadrić M, Brianti E, Duscher GG (2016) Red fox (Vulpes vulpes) as a potential reservoir host of cardiorespiratory parasites in Bosnia and Herzegovina. Vet Parasitol 223:63–70
Jahren T, Odden M, Linnell JD, Panzacchi M (2020) The impact of human land use and landscape productivity on population dynamics of red fox in southeastern Norway. Mamm Res 65:503–516
Jahren T (2017) The role of nest predation and nest predators in population declines of capercaillie and black grouse. PhD dissertation, Faculty of applied ecology and agricultural sciences, Inland Norway University of Applied Sciences, Evenstad, Norway
Jiménez J, Chandler R, Tobajas J, Descalzo E, Mateo R, Ferreras P (2019) Generalized spatial mark–resight models with incomplete identification: An application to red fox density estimates. Ecol Evol 9(8):4739–4748
Jiménez J, Augustine C, B, Linden DW, Chandler RB, Royle JA, (2021) Spatial capture–recapture with random thinning for unidentified encounters. Ecol Evol 11(3):1187–1198
Kämmerle JL, Corlatti L, Harms L, Storch I (2018) Methods for assessing small-scale variation in the abundance of a generalist mesopredator. PLoS ONE 13(11):e0207545
Kellner KF, Swihart RK (2014) Accounting for imperfect detection in ecology: a quantitative review. PLoS ONE 9(10):e111436
Kery M, Gardner B, Stoeckle T, Weber D, Royle JA (2011) Use of spatial capture–recapture modeling and DNA data to estimate densities of elusive animals. Conserv Biol 25(2):356–364
Killengreen ST, Lecomte N, Ehrich D, Schott T, Yoccoz NG, Ims RA (2011) The importance of marine vs. human‐induced subsidies in the maintenance of an expanding mesocarnivore in the arctic tundra. J Animal Ecol 80(5): 1049–1060
Larivière S, Pasitschniak-Arts M (1996) Vulpes vulpes. Mamm Spec 537:1–11
Laurimaa L, Moks E, Soe E, Valdmann H, Saarma U (2016) Echinococcus multilocularis and other zoonotic parasites in red foxes in Estonia. Parasitology 143(11):1450
Lindström E (1989) Food limitation and social regulation in a red fox population. Ecography 12(1):70–79
Lindström ER, Andrén H, Angelstam P, Cederlund G, Hörnfeldt B, Jäderberg L, Lemnell PA, Martinsson B, Sköld K, Swenson JE (1994) Disease reveals the predator: sarcoptic mange, red fox predation, and prey populations. Ecology 75(4):1042–1049
Milleret C, Dupont P, Chipperfield J, Turek D, Brøseth H, Gimenez O, de Valpine P, Bischof R (2020) Estimating abundance with interruptions in data collection using open population spatial capture–recapture models. Ecosphere 11(7): e03172.
Miquel C, Bellemain E, Poillot C, Bessiere A, Durand A, Taberlet P (2006) Quality indexes to assess the reliability of genotypes in studies using non-invasive sampling and multiple-tube approach. Mol Ecol Notes 6:985–988
Moen A (1998) National Atlas of Norway: Vegetation (Original title: Vegetasjonsatlas for Norge: vegetasjon). Norwegian Mapping Authority, Hønefoss.
Molina S, Fuller AK, Morin DJ, Royle JA (2017) Use of spatial capture–recapture to estimate density of Andean bears in northern Ecuador. Ursus 28(1):117–126
Moore M, Brown SK, Sacks BN (2010a) Thirty-one short red fox (Vulpes vulpes) microsatellite markers. Mol Ecol Resour 10:404–408
Moore SM, Borer ET, Hosseini PR (2010b) Predators indirectly control vector-borne disease: linking predator–prey and host–pathogen models. J R Soc Interface 7(42):161–176
Morin DJ, Kelly MJ, Waits LP (2016) Monitoring coyote population dynamics with fecal DNA and spatial capture–recapture. J Wildl Manag 80(5):824–836
Mumma MA, Zieminski C, Fuller TK, Mahoney SP, Waits LP (2015) Evaluating noninvasive genetic sampling techniques to estimate large carnivore abundance. Mol Ecol Resour 15(5):1133–1144
Needham R, Odden M, Lundstadsveen SK, Wegge P (2014) Seasonal diets of red foxes in a boreal forest with a dense population of moose: the importance of winter scavenging. Acta Theriol 59(3):391–398
Norwegian Mapping Authority (2020) N50 Kartdata. April 2020, URL: https://register.geonorge.no/det-offentlige-kartgrunnlaget/n50-kartdata/ea192681-d039-42ec-b1bc-f3ce04c189ac
O’Connell AF, Nichols JD, Karanth KU (eds) (2010) Camera traps in animal ecology: methods and analyses. Springer Science and Business Media, Berlin
Østbye E, Steen H, Framstad E, Tvete B (1989) Do connections exist between climatic variations and cyclicity in small rodents? Fauna 42:147–153
Panasci M, Ballard WB, Breck S, Rodriguez D, Densmore LD III, Wester DB, Baker RJ (2011) Evaluation of fecal DNA preservation techniques and effects of sample age and diet on genotyping success. J Wildl Manag 75(7):1616–1624
Piggott MP (2005) Effect of sample age and season of collection on the reliability of microsatellite genotyping of faecal DNA. Wildl Res 31(5):485–493
QGIS Development Team (2019) QGIS Geographic Information System. Open Source Geospatial Foundation Project. URL: http://qgis.osgeo.org
R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: https://www.R-project.org/
Rød-Eriksen L, Skrutvold J, Herfindal I, Jensen H, Eide NE (2020) Highways associated with expansion of boreal scavengers into the alpine tundra of Fennoscandia. J Appl Ecol 57:1861–1870
Royle JA, Young KV (2008) A hierarchical model for spatial capture–recapture data. Ecology 89(8):2281–2289
Royle JA, Chandler RB, Sollmann R, Gardner B (2013) Spatial capture–recapture. Academic Press, Cambridge
Royle JA, Fuller AK, Sutherland C (2018) Unifying population and landscape ecology with spatial capture–recapture. Ecography 41(3):444–456
Silvy NJ (Eds.) (2012) The Wildlife Techniques Manual: Volume 1: Research. Volume 2: Management 2-vol. Set (Vol. 1). JHU Press, Baltimore.
Skrede A (2016) Nest-predation in willow ptarmigan – Red fox in the mountains (Original title: Reirpredasjon hjå lirype–Raudrev inntar fjellet). Bachelor's thesis, Faculty of Applied Ecology and Agricultural Sciences, Inland Norway University of Applied Sciences, Evenstad, Norway
Smedshaug CA, Selås V, Lund SE, Sonerud GA (1999) The effect of a natural reduction of red fox Vulpes vulpes on small game hunting bags in Norway. Wildl Biol 5(1):157–166
Sollmann R, Mohamed A, Samejima H, Wilting A (2013) Risky business or simple solution–Relative abundance indices from camera-trapping. Biol Cons 159:405–412
Statistics Norway (2020) Population, by sex and one-year age groups. Read 27. April 2020, URL: https://www.ssb.no/en/befolkning/statistikker/folkemengde
Sun CC, Fuller AK, Royle JA (2014) Trap configuration and spacing influences parameter estimates in spatial capture–recapture models. PLoS ONE 9(2):e88025
Sutherland C, Royle JA, Linden DW (2019) oSCR: a spatial capture–recapture R package for inference about spatial ecological processes. Ecography 42(9):1459–1469
Svendsen K (2016) Movement and habitat use of red foxes in a boreal conifer forest (Original title: Rødrevens bevegelsesmønster og habitatbruk i en boreal barskog). Bachelor's thesis, Faculty of Applied Ecology and Agricultural Sciences, Inland Norway University of Applied Sciences, Evenstad
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24(16):3189–3194
Tourani M, Dupont P, Nawaz MA, Bischof R (2020) Multiple observation processes in spatial capture–recapture models: How much do we gain? Ecology 101(7): e03030.
Van Etten KW, Wilson KR, Crabtree RL (2007) Habitat use of red foxes in Yellowstone National Park based on snow tracking and telemetry. J Mammal 88(6):1498–1507
Víchová B, Bona M, Miterpáková M, Kraljik J, Čabanová V, Nemčíková G, Hurníková Z, Oravec M (2018) Fleas and ticks of red foxes as vectors of canine bacterial and parasitic pathogens, in Slovakia. Central Europe Vector-Borne and Zoonotic Diseases 18(11):611–619
Vine SJ, Crowther MS, Lapidge SJ, Dickman CR, Mooney N, Piggott MP, English AW (2009) Comparison of methods to detect rare and cryptic species: a case study using the red fox (Vulpes vulpes). Wildl Res 36(5):436–446
Walton Z, Mattisson J (2021) Down a hole: missing GPS positions reveal birth dates of an underground denning species, the red fox. Mamm Biol 101(3):357–362
Walton Z, Samelius G, Odden M, Willebrand T (2017) Variation in home range size of red foxes Vulpes vulpes along a gradient of productivity and human landscape alteration. PLoS ONE 12(4):e0175291
Webbon CC, Baker PJ, Harris S (2004) Faecal density counts for monitoring changes in red fox numbers in rural Britain. J Appl Ecol 41(4):768–779
Wegge P, Rolstad J (2011) Clearcutting forestry and Eurasian boreal forest grouse: long-term monitoring of sympatric capercaillie Tetrao urogallus and black grouse T tetrix reveals unexpected effects on their population performances. Forest Ecol Manag 261(9):1520–1529
Wegge P, Bakke BB, Odden M, Rolstad J (2019) DNA from scats combined with capture–recapture modeling: a promising tool for estimating the density of red foxes—a pilot study in a boreal forest in southeast Norway. Mamm Res 64(1):147–154
Woodruff SP, Johnson TR, Waits LP (2015) Evaluating the interaction of faecal pellet deposition rates and DNA degradation rates to optimize sampling design for DNA-based mark–recapture analysis of Sonoran pronghorn. Mol Ecol Resour 15(4):843–854
Acknowledgements
We thank all field personnel, students, and volunteers who were involved in collecting samples in the field. The project “How many foxes are out there?” was funded by the Norwegian Environmental Agency (contract 18S247E2) and several grants from the County Governors and County Municipalites in Trøndelag, Møre-Romsdal, Viken, Vestfold, and Telemark. The project builds into the ECOFUNC project funded by the Research Council of Norway (grant 244557/E50), as well as project WildMap (NFR 286886).
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital).
Author information
Authors and Affiliations
Contributions
NEE, LRE, and ØF conceived the research idea. NEE, KRU, LRE, and LKL coordinated the field work. IPØA and LKL performed the genetic analyses. LKL, PD, and RB designed and performed the SCR analyses. LKL wrote the manuscript, and all authors contributed critically to the draft.
Corresponding author
Additional information
Communicated by Christian Kiffner.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lindsø, L.K., Dupont, P., Rød-Eriksen, L. et al. Estimating red fox density using non-invasive genetic sampling and spatial capture–recapture modelling. Oecologia 198, 139–151 (2022). https://doi.org/10.1007/s00442-021-05087-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-021-05087-3