Abstract
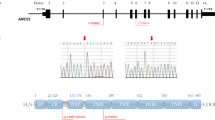
Precision medicine requires precise genetic variant interpretation, yet many disease-associated genes have unresolved variants of unknown significance (VUS). We analyzed variants in a well-studied gene, FGFR1, a common cause of Idiopathic Hypogonadotropic Hypogonadism (IHH) and examined whether regional genetic enrichment of missense variants could improve variant classification. FGFR1 rare sequence variants (RSVs) were examined in a large cohort to (i) define regional genetic enrichment, (ii) determine pathogenicity based on the American College of Medical Genetics/Association for Molecular Pathology (ACMG/AMP) variant classification framework, and (iii) characterize the phenotype of FGFR1 variant carriers by variant classification. A total of 143 FGFR1 RSVs were identified in 175 IHH probands (n = 95 missense, n = 48 protein-truncating variants). FGFR1 missense RSVs showed regional enrichment across biologically well-defined domains: D1, D2, D3, and TK domains and linker regions (D2–D3, TM–TK). Using these defined regions of enrichment to augment the ACMG/AMP classification reclassifies 37% (20/54) of FGFR1 missense VUS as pathogenic or likely pathogenic (PLP). Non-proband carriers of FGFR1 missense VUS variants that were reclassified as PLP were more likely to express IHH or IHH-associated phenotypes [anosmia or delayed puberty] than non-proband carriers of FGFR1 missense variants that remained as VUS (76.9% vs 34.7%, p = 0.035). Using the largest cohort of FGFR1 variant carriers, we show that integration of regional genetic enrichment as moderate evidence for pathogenicity improves the classification of VUS and that reclassified variants correlated with phenotypic expressivity. The addition of regional genetic enrichment to the ACMG/AMP guidelines may improve clinical variant interpretation.



Similar content being viewed by others
Data Availability
Confirmed sequencing data for variants in this manuscript are deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/; submissions SCV003932463–SCV003932605). Data generated or analyzed during this study are included in this published article and its supplementary information files. Additional data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. https://doi.org/10.1002/0471142905.hg0720s76
Balasubramanian R, Crowley WF Jr (2011) Isolated GnRH deficiency: a disease model serving as a unique prism into the systems biology of the GnRH neuronal network. Mol Cell Endocrinol 346(1–2):4–12. https://doi.org/10.1016/j.mce.2011.07.012
Belov AA, Mohammadi M (2013) Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a015958
Brnich SE, Abou Tayoun AN, Couch FJ et al (2019) Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 12(1):3. https://doi.org/10.1186/s13073-019-0690-2
Burke W, Parens E, Chung WK, Berger SM, Appelbaum PS (2022) The challenge of genetic variants of uncertain clinical significance : a narrative review. Ann Intern Med 175(7):994–1000. https://doi.org/10.7326/M21-4109
Cangiano B, Swee DS, Quinton R, Bonomi M (2021) Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease. Hum Genet 140(1):77–111. https://doi.org/10.1007/s00439-020-02147-1
Chora JR, Iacocca MA, Tichy L et al (2022) The Clinical Genome Resource (ClinGen) familial hypercholesterolemia variant curation expert panel consensus guidelines for LDLR variant classification. Genet Med 24(2):293–306. https://doi.org/10.1016/j.gim.2021.09.012
Dode C, Levilliers J, Dupont JM et al (2003) Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet 33(4):463–465. https://doi.org/10.1038/ng1122
Doty RL, Shaman P, Dann M (1984) Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol Behav 32(3):489–502. http://www.ncbi.nlm.nih.gov/pubmed/6463130
Doty RL, Marcus A, Lee WW (1996) Development of the 12-item Cross-Cultural Smell Identification Test (CC-SIT). The Laryngoscope 106(3 Pt 1):353–6. http://www.ncbi.nlm.nih.gov/pubmed/8614203
Gelman H, Dines JN, Berg J et al (2019) Recommendations for the collection and use of multiplexed functional data for clinical variant interpretation. Genome Med 11(1):85. https://doi.org/10.1186/s13073-019-0698-7
Genomes Project C, Auton A, Brooks LD et al (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF (2018) Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet 103(4):522–534. https://doi.org/10.1016/j.ajhg.2018.08.016
Harrison SM, Biesecker LG, Rehm HL (2019) Overview of specifications to the ACMG/AMP variant interpretation guidelines. Curr Protoc Hum Genet 103(1):e93. https://doi.org/10.1002/cphg.93
Ioannidis NM, Rothstein JH, Pejaver V et al (2016) REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 99(4):877–885. https://doi.org/10.1016/j.ajhg.2016.08.016
Kallmann FJ, Schönfeld WA, Barrera SE (1944) The genetic aspects of primary eunuchoidism. Am J Ment Def 48:203–236
Kelly MA, Caleshu C, Morales A et al (2018) Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med 20(3):351–359. https://doi.org/10.1038/gim.2017.218
Kim SH, Hu Y, Cadman S, Bouloux P (2008) Diversity in fibroblast growth factor receptor 1 regulation: learning from the investigation of Kallmann syndrome. J Neuroendocrinol 20(2):141–163. https://doi.org/10.1111/j.1365-2826.2007.01627.x
Lansdon LA, Bernabe HV, Nidey N, Standley J, Schnieders MJ, Murray JC (2017) The use of variant maps to explore domain-specific mutations of FGFR1. J Dent Res 96(11):1339–1345. https://doi.org/10.1177/0022034517726496
Lee K, Krempely K, Roberts ME et al (2018) Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants. Hum Mutat 39(11):1553–1568. https://doi.org/10.1002/humu.23650
Lek M, Karczewski KJ, Minikel EV et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. https://doi.org/10.1038/nature19057
Lewkowitz-Shpuntoff HM, Hughes VA, Plummer L et al (2012) Olfactory phenotypic spectrum in idiopathic hypogonadotropic hypogonadism: pathophysiological and genetic implications. J Clin Endocrinol Metab 97(1):E136–E144. https://doi.org/10.1210/jc.2011-2041
McKenna A, Hanna M, Banks E et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31(13):3812–3814. https://doi.org/10.1093/nar/gkg509
O’Connor TD, Kiezun A, Bamshad M et al (2013) Fine-scale patterns of population stratification confound rare variant association tests. PLoS ONE 8(7):e65834. https://doi.org/10.1371/journal.pone.0065834
Patel MJ, DiStefano MT, Oza AM et al (2021) Disease-specific ACMG/AMP guidelines improve sequence variant interpretation for hearing loss. Genet Med 23(11):2208–2212. https://doi.org/10.1038/s41436-021-01254-2
Pitteloud N, Meysing A, Quinton R et al (2006) Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol 254–255:60–69. https://doi.org/10.1016/j.mce.2006.04.021
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Ross JE, Zhang BM, Lee K et al (2021) Specifications of the variant curation guidelines for ITGA2B/ITGB3: ClinGen Platelet Disorder Variant Curation Panel. Blood Adv 5(2):414–431. https://doi.org/10.1182/bloodadvances.2020003712
Samocha KE, Kosmicki JA, Karczewski KJ et al (2017) Regional missense constraint improves variant deleteriousness prediction. bioRxiv. https://doi.org/10.1101/148353
Shirts BH, Pritchard CC, Walsh T (2016) Family-specific variants and the limits of human genetics. Trends Mol Med 22(11):925–934. https://doi.org/10.1016/j.molmed.2016.09.007
Tian Y, Pesaran T, Chamberlin A et al (2019) REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci Rep 9(1):12752. https://doi.org/10.1038/s41598-019-49224-8
Topaloglu AK (2017) Update on the genetics of idiopathic hypogonadotropic hypogonadism. J Clin Res Pediatr Endocrinol 9(Suppl 2):113–122. https://doi.org/10.4274/jcrpe.2017.S010
Villanueva C, Jacobson-Dickman E, Xu C et al (2015) Congenital hypogonadotropic hypogonadism with split hand/foot malformation: a clinical entity with a high frequency of FGFR1 mutations. Genet Med 17(8):651–659. https://doi.org/10.1038/gim.2014.166
Walsh R, Mazzarotto F, Whiffin N et al (2019) Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med 11(1):5. https://doi.org/10.1186/s13073-019-0616-z
Acknowledgements
This work was supported by the Eunice K. Shriver National Institute for Child Health and Human Development (P50 HD-28138 to S.B.S., support for W.X. and L.P., 1K23HD097296 to M.F.L., K23 HD077043 to R.B., R01 HD096324 to R.B.); Catalyst Medical Research Investigator Training Award to M.F.L.; National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541 and by financial contributions from Harvard University and its affiliated academic health care centers to Harvard Catalyst/the Harvard Clinical and Translational Science Center. S.B.S. is a Robert and Laura Reynolds Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, its affiliated academic health care centers, or the National Institutes of Health. We thank the research subjects and referring providers [Fotini Adamidou, Tania Aguirre, Iram Ahmad, Abdulmoein Al-Agha, Jonathan Anolik, Anita Azam, Bert Bachrach, Hagit Baris, Gerhard Baumann, Lynn Bennion, Pierre Bouloux, Jan Bruder, Jean Pierre Chanoine, Jeannette Chinchilla, Jin-Ho Choi, Thomas P Clairmont, Amy Criego, Ashwin Dalal, Angela Delaney, Rachel Edelen, Deborah Elder, Micheal Fili, Eric Fliers, John Fowlkes, Larry Fox, Peter Fuller, Dieter Furthner, Neoklis Georgopoulos, Nadine Haddad, Sheri Horsburgh, Louise Izatt, Silvia Kaufmann, Sjoberg Kho, Susan Kirsch, Peter Kopp, Mariarosaria Lang-Muritano, Helene Lavoie, Peter Lee, Selwyn B. Levitt, Irene Mamkin, Veronica Mericq, Daniel Metzger, Kiyonori Miura, Ron Newfield, Metin Ozata, Jose Perez-Rodiguez, Duarte Pignatelli, Richard Quintin, Jose Quintos, Sally Radovick, Cesar Ramos-Remus, Barry Reiner, Elise Rodman, Craig Sadur, Nicole Simon, Arnold Slyper, Rebecca Sokol, Susan Sparks, Phyllis Speiser, Dennis Styne, Diane Suchet, Akira Takeshita, Arthi Thirumalai, Sherida Tollefsen, Kemal Topalolgu, Marshall Tulloch-Reid, Guy Van Vliet, Charles Verge, Teresa Vieira, Darren West, Melissa Woo], without whom this research would not be possible. We also thank Kathy Salnikov and other members of the MGH Reproductive Endocrine Unit for their assistance and input. We thank Andrew Dwyer for reviewing the manuscript. We thank the staff of the Broad Genomic Core.
Funding
This work was supported by the Eunice K. Shriver National Institute for Child Health and Human Development (P50 HD-28138 to S.B.S., support for W.X. and L.P., 1K23HD097296 to M.F.L., K23 HD077043 to R.B., R01 HD096324 to R.B.); Catalyst Medical Research Investigator Training Award to M.F.L.; National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541 and by financial contributions from Harvard University and its affiliated academic health care centers to Harvard Catalyst/the Harvard Clinical and Translational Science Center. S.B.S. is a Robert and Laura Reynolds Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, its affiliated academic health care centers, or the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
WX and MFL contributed to the study conception and design. Material preparation, data collection and analysis were performed by WX, MFL, LP. Data interpretation was performed by WX, MFL, RB, SBS. The first draft of the manuscript was written by WX and MFL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. This research was reviewed and approved by the Massachusetts General Hospital Partners Institutional Review Board (ClinicalTrials.gov no. NCT00494169; registered 29 June 2007).
Consent to participate
Written informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, W., Plummer, L., Seminara, S.B. et al. How human genetic context can inform pathogenicity classification: FGFR1 variation in idiopathic hypogonadotropic hypogonadism. Hum. Genet. 142, 1611–1619 (2023). https://doi.org/10.1007/s00439-023-02601-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02601-w