Abstract
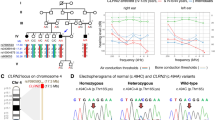
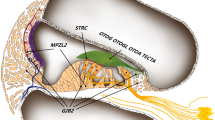
Hereditary hearing impairment is a common sensory disorder that is genetically and phenotypically heterogeneous. In this study, we used a homozygosity mapping and exome sequencing strategy to study a consanguineous Pakistani family with autosomal recessive severe-to-profound hearing impairment. This led to the identification of a missense variant (p.Ile369Thr) in the LMX1A gene affecting a conserved residue in the C-terminus of the protein, which was predicted damaging by an in silico bioinformatics analysis. The p.Ile369Thr variant disrupts several C-terminal and homeodomain residue interactions, including an interaction with homeodomain residue p.Val241 that was previously found to be involved in autosomal dominant progressive HI. LIM-homeodomain factor Lmx1a is expressed in the inner ear through development, shows a progressive restriction to non-sensory epithelia, and is important in the separation of the sensory and non-sensory domains in the inner ear. Homozygous Lmx1a mutant mice (Dreher) are deaf with dysmorphic ears with an abnormal morphogenesis and fused and misshapen sensory organs; however, computed tomography performed on a hearing-impaired family member did not reveal any cochleovestibular malformations. Our results suggest that LMX1A is involved in both human autosomal recessive and dominant sensorineural hearing impairment.


Similar content being viewed by others
References
Ahmed ZM, Riazuddin S, Ahmad J et al (2003) PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet 12:3215–3223. https://doi.org/10.1093/hmg/ddg358
Alagramam KN, Stahl JS, Jones SM et al (2005) Characterization of vestibular dysfunction in the mouse model for usher syndrome 1F. J Assoc Res Otolaryngol 6:106–118. https://doi.org/10.1007/s10162-004-5032-3
Alharazneh A, Luk L, Huth M et al (2011) Functional hair cell mechanotransducer channels are required for aminoglycoside ototoxicity. PLoS One 6:e22347. https://doi.org/10.1371/journal.pone.0022347
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aoki K, Taketo MM (2007) Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci 120:3327–3335. https://doi.org/10.1242/jcs.03485
Auton A, Abecasis GR, Altshuler DM et al (2015) A global reference for human genetic variation. Nature 526:68–74. https://doi.org/10.1038/nature15393
Bateman A, Martin MJ, O’Donovan C et al (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. https://doi.org/10.1093/nar/gkw1099
Bhati M, Lee C, Nancarrow AL et al (2008) Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J 27:2018–2029. https://doi.org/10.1038/emboj.2008.123
Chizhikov V, Steshina E, Roberts R et al (2006) Molecular definition of an allelic series of mutations disrupting the mouse Lmx1a (dreher) gene. Mamm Genome 17:1025–1032. https://doi.org/10.1007/s00335-006-0033-7
del Castillo FJ, del Castillo I (2017) DFNB1 non-syndromic hearing impairment: diversity of mutations and associated phenotypes. Front Mol Neurosci 10:428. https://doi.org/10.3389/fnmol.2017.00428
Doucet-Beaupré H, Ang S-L, Lévesque M (2015) Cell fate determination, neuronal maintenance and disease state: the emerging role of transcription factors Lmx1a and Lmx1b. FEBS Lett 589:3727–3738. https://doi.org/10.1016/j.febslet.2015.10.020
Dreyer SD, Morello R, German MS et al (2000) LMX1B transactivation and expression in nail-patella syndrome. Hum Mol Genet 9:1067–1074. https://doi.org/10.1093/hmg/9.7.1067
Dunston JA, Hamlington JD, Zaveri J et al (2004) The human LMX1B gene: transcription unit, promoter, and pathogenic mutations. Genomics 84:565–576. https://doi.org/10.1016/J.YGENO.2004.06.002
Elahi MM, Elahi F, Elahi A, Elahi SB (1998) Paediatric hearing loss in rural Pakistan. J Otolaryngol 27:348–353
Eswar N, Webb B, Marti-Renom MA et al (2007) Comparative protein structure modeling using MODELLER. In: Current protocols in protein science. Wiley, Hoboken, 2.9.1–2.9.31
Failli V, Bachy I, Rétaux S (2002) Expression of the LIM-homeodomain gene Lmx1a (dreher) during development of the mouse nervous system. Mech Dev 118:225–228
Fu W, O’Connor TD, Jun G et al (2012) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–220. https://doi.org/10.1038/nature11690
Glover JC, Elliott KL, Erives A et al (2018) Wilhelm His’ lasting insights into hindbrain and cranial ganglia development and evolution. Dev Biol. https://doi.org/10.1016/j.ydbio.2018.02.001 (in Press)
Hedlund E, Belnoue L, Theofilopoulos S et al (2016) Dopamine receptor antagonists enhance proliferation and neurogenesis of midbrain Lmx1a-expressing progenitors. Sci Rep 6:26448. https://doi.org/10.1038/srep26448
Hussain R, Bittles AH (1998) The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci 30:261–275
Kircher M, Witten DM, Jain P et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. https://doi.org/10.1038/ng.2892
Koo SK, Hill JK, Hwang CH et al (2009) Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol 333:14–25. https://doi.org/10.1016/j.ydbio.2009.06.016
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Landrum MJ, Lee JM, Benson M et al (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44:D862–D868. https://doi.org/10.1093/nar/gkv1222
Lek M, Karczewski KJ, Minikel EV et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. https://doi.org/10.1038/nature19057
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Liu XZ, Hope C, Walsh J et al (1998) Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet 63:909–912. https://doi.org/10.1086/302026
Liu X, Jian X, Boerwinkle E (2011) dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32:894–899. https://doi.org/10.1002/humu.21517
Mann ZF, Gálvez H, Pedreno D et al (2017) Shaping of inner ear sensory organs through antagonistic interactions between Notch signalling and Lmx1a. Elife 6:e33323. https://doi.org/10.7554/eLife.33323
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Nichols DH, Pauley S, Jahan I et al (2008) Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res 334:339–358. https://doi.org/10.1007/s00441-008-0709-2
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Riazuddin S, Belyantseva IA, Giese APJ et al (2012) Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet 44:1265–1271. https://doi.org/10.1038/ng.2426
Ritter N, Kilinc E, Navruz B, Bae Y (2011) Test Review: L. Brown, R. J. Sherbenou, & S. K. Johnsen test of nonverbal intelligence-4 (TONI-4). Austin, TX: PRO-ED,2010. J Psychoeduc Assess 29:484–488. https://doi.org/10.1177/0734282911400400
Sambrook J, Russell DW (2006) Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 2006:pdb.prot4455. https://doi.org/10.1101/pdb.prot4455
Schultz JM, Khan SN, Ahmed ZM et al (2009) Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet 85:25–39. https://doi.org/10.1016/j.ajhg.2009.06.003
Scott EM, Halees A, Itan Y et al (2016) Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48:1071–1076. https://doi.org/10.1038/ng.3592
Seelow D, Schuelke M (2012) HomozygosityMapper2012-bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res 40:W516–W520. https://doi.org/10.1093/nar/gks487
Shahzad M, Sivakumaran TA, Qaiser TA et al (2013) Genetic analysis through OtoSeq of Pakistani families segregating prelingual hearing loss. Otolaryngol Neck Surg 149:478–487. https://doi.org/10.1177/0194599813493075
Shearer AE, Smith RJH (2012) Genetics: advances in genetic testing for deafness. Curr Opin Pediatr 24:679–686. https://doi.org/10.1097/MOP.0b013e3283588f5e
Steffes G, Lorente-Cánovas B, Pearson S et al (2012) Mutanlallemand (mtl) and belly spot and deafness (bsd) are two new mutations of Lmx1a causing severe cochlear and vestibular defects. PLoS One 7:e51065. https://doi.org/10.1371/journal.pone.0051065
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164–e164. https://doi.org/10.1093/nar/gkq603
Wesdorp M, de Koning Gans PAM, Schraders M et al (2018) Heterozygous missense variants of LMX1A lead to nonsyndromic hearing impairment and vestibular dysfunction. Hum Genet. https://doi.org/10.1007/s00439-018-1880-5
Acknowledgements
This work was supported by the Higher Education Commission of Pakistan (to WA) and the National Institute on Deafness and Other Communication Disorders grants R01 DC011651 and R01 DC003594 (to SML). Exome sequencing was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant HG006493 (to DAN, MJB, and SML). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I. We would like to acknowledge the entire University of Washington Center for Mendelian Genomics team and their members for exome sequencing and bioinformatics support: Michael J. Bamshad (University of Washington, Seattle Children’s Hospital), Suzanne M. Leal (Baylor College of Medicine), and Deborah A. Nickerson (University of Washington); Peter Anderson (University of Washington), Marcus Annable (University of Washington), Elizabeth E. Blue (University of Washington), Kati J. Buckingham (University of Washington), Imen Chakchouk (Baylor College of Medicine), Jennifer Chin (University of Washington), Jessica X Chong (University of Washington), Rodolfo Cornejo Jr. (University of Washington), Colleen P. Davis (University of Washington), Christopher Frazar (University of Washington), Martha Horike-Pyne (University of Washington), Gail P. Jarvik (University of Washington), Eric Johanson (University of Washington), Ashley N. Kang (University of Washington), Tom Kolar (University of Washington), Stephanie A. Krauter (University of Washington), Colby T. Marvin (University of Washington), Sean McGee (University of Washington), Daniel J. McGoldrick (University of Washington), Karynne Patterson (University of Washington), Sam W. Phillips (University of Washington), Jessica Pijoan (University of Washington), Matthew A. Richardson (University of Washington), Peggy D. Robertson (University of Washington), Isabelle Schrauwen (Baylor College of Medicine), Krystal Slattery (University of Washington), Kathryn M. Shively (University of Washington), Joshua D. Smith (University of Washington), Monica Tackett (University of Washington), Alice E. Tattersall (University of Washington), Marc Wegener (University of Washington), Jeffrey M. Weiss (University of Washington), Marsha M. Wheeler (University of Washington), Qian Yi (University of Washington), and Di Zhang (Baylor College of Medicine).
Online weblinks: ANNOVAR, http://annovar.openbioinformatics.org/, BLASTP, https://blast.ncbi.nlm.nih.gov/Blast.cgi/, Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/, Combined Annotation-Dependent Depletion (CADD), http://cadd.gs.washington.edu/, dbNSFP, https://sites.google.com/site/jpopgen/dbNSFP, dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/, Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/, Genome Analysis Toolkit (GATK), https://software.broadinstitute.org/gatk/, Greater Middle East (GME) Variome Project, http://igm.ucsd.edu/gme, International Organization for Standardization (ISO), https://www.iso.org/, MODELLER8v1 software, https://salilab.org/modeller/, Multiple Alignment tool, http://www.bioinformatics.org/sms/multi_align.html, Picard, http://broadinstitute.github.io/picard/, UCSF Chimera, https://www.cgl.ucsf.edu/chimera/, and Universal Protein Resource (UniProt), http://www.uniprot.org/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2018_1899_MOESM2_ESM.xlsx
ANNOVAR and dbNSFP annotations of the variant in LMX1A and all additional rare variants of unknown significance identified by exome sequencing in individual IV:2 (XLSX 16 KB)
Rights and permissions
About this article
Cite this article
Schrauwen, I., Chakchouk, I., Liaqat, K. et al. A variant in LMX1A causes autosomal recessive severe-to-profound hearing impairment. Hum Genet 137, 471–478 (2018). https://doi.org/10.1007/s00439-018-1899-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-018-1899-7