Abstract
Uterine fibroids are benign tumors of the uterus affecting up to 77% of women by menopause. They are the leading indication for hysterectomy, and account for $34 billion annually in the United States. Race/ethnicity and age are the strongest known risk factors. African American (AA) women have higher prevalence, earlier onset, and larger and more numerous fibroids than European American women. We conducted a multi-stage genome-wide association study (GWAS) of fibroid risk among AA women followed by in silico genetically predicted gene expression profiling of top hits. In Stage 1, cases and controls were confirmed by pelvic imaging, genotyped and imputed to 1000 Genomes. Stage 2 used self-reported fibroid and GWAS data from 23andMe, Inc. and the Black Women’s Health Study. Associations with fibroid risk were modeled using logistic regression adjusted for principal components, followed by meta-analysis of results. We observed a significant association among 3399 AA cases and 4764 AA controls at rs739187 (risk-allele frequency = 0.27) in CYTH4 (OR (95% confidence interval) = 1.23 (1.16–1.30), p value = 7.82 × 10−9). Evaluation of the genetic association results with MetaXcan identified lower predicted gene expression of CYTH4 in thyroid tissue as significantly associated with fibroid risk (p value = 5.86 × 10−8). In this first multi-stage GWAS for fibroids among AA women, we identified a novel risk locus for fibroids within CYTH4 that impacts gene expression in thyroid and has potential biological relevance for fibroids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Uterine fibroids or leiomyomata are common, benign tumors of the uterus with an estimated lifetime risk of 77% by menopause (Baird et al. 2003). African Americans (AA) are more likely to have fibroids than women of European ancestry (EA), with AA having greater than 80% incidence of fibroids by menopause compared to nearly 70% for EA (Baird et al. 2003). AA women also have larger and more numerous fibroids as well as a younger age-of-onset on average (Baird et al. 2003). In addition to race/ethnicity (Baird et al. 2003; Faerstein et al. 2001; Marshall et al. 1997), there are other well-characterized risk factors for fibroids, including early age at menarche (Baird and Dunson 2003; Cha et al. 2011; Faerstein et al. 2001; Luoto et al. 2000; Moore et al. 2008), being overweight (BMI 25–29 kg/m2) (Baird et al. 2007; Moore et al. 2008; Terry et al. 2007; Wise et al. 2005a), and older premenopausal age (Baird et al. 2003; Moore et al. 2008). In addition, higher parity is associated with reduced fibroid risk, likely due to pregnancy-related hormonal and uterine changes (Baird and Dunson 2003). Symptoms of uterine fibroids may include pelvic pain and abnormal or heavy menses, though many fibroids are asymptomatic (Baird et al. 2003; Borah et al. 2013; Vollenhoven 1998). The lack of overt symptoms makes imaging crucial for classification of case/control status, as up to 51% of women may be misclassified by self-report (Baird et al. 2003; Myers et al. 2012). We have developed and validated a phenotyping algorithm to classify fibroid case/control status using electronic health records (Feingold-Link et al. 2014). This algorithm requires pelvic imaging for identification of cases and controls, which reduces misclassification of both cases and controls.
Several lines of evidence suggest genetic risk factors influence fibroid development. Estimates of the heritability of fibroids from familial aggregation and twin studies range from 26 to 69% in European populations (Luoto et al. 2000). Racial disparities in age at onset, number, size, and lifetime incidence of fibroids by menopause (Baird et al. 2003) also strongly support a role for genetic factors. There has been one published genome-wide association study (GWAS) of uterine fibroids, which was performed among Japanese subjects (Cha et al. 2011). Genome-wide linkage and follow-up association studies in a meta-analysis of EA women implicated an additional locus for risk of fibroid diagnosis (Eggert et al. 2012). The loci implicated in these previous studies (SLK, BET1, TNRC6B, and FASN/CCDC57) have been replicated among EAs (Aissani et al. 2015a; Edwards et al. 2013b), which have been evaluated but failed to replicate in AAs (Aissani et al. 2015b; Wise et al. 2012). The predominant studies conducted among AA subjects have involved admixture mapping, which has shown significant regions of increased African ancestry in cases, particularly at 1q42.2 (Zhang et al. 2015), 4p16, and 10q26 (Wise et al. 2012). The aim of this work was to perform the first GWAS for fibroids risk among AA women using image-verified fibroids for discovery, with replication in cohorts that collected data on self-reported fibroids.
Methods
Study populations
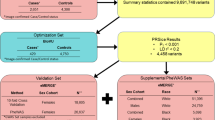
Individuals with imaging-confirmed uterine fibroids and genome-wide genotype data were included from the following studies in Stage 1: Vanderbilt University BioVU, Mt Sinai, BioME and the Coronary Artery Risk Development in Young Adults (CARDIA) Women’s Study (CARDIA-WS) for a total of 1273 cases and 1379 controls. All studies received Institutional Review Board approval at their respective institutions and written informed consent was obtained for all participants.
BioVU is an electronic health record (EHR) biorepository at Vanderbilt University Medical Center, Nashville, TN and was designed to link clinical data available from de-identified EHRs to DNA specimens. Methods have been previously described (Pulley et al. 2010). BioME is the ongoing, consented EHR-linked biobank at the Institute for Personalized Medicine at the Icahn School of Medicine at Mt. Sinai. A subset of the available BioME samples were genotyped as part of the Electronic Medical Records and Genetics (eMERGE) Network and are referred to in this instance as Mt. Sinai, while genotyped samples acquired through other means were analyzed separately and will be henceforth referred to as BioME. The phenotyping algorithms used to identify cases and controls in BioVU, Mt. Sinai, and BioME have been previously published (Feingold-Link et al. 2014). Briefly, the phenotyping algorithm required at least one instance of pelvic imaging with a diagnosis code for fibroids to define cases among women aged 18 and over. For controls, at least two instances of pelvic imaging were required, on different dates, with no diagnosis or procedure codes indicating a fibroid, fibroid treatment, or hysterectomy. The CARDIA study is a prospective multi-center study with 5115 adult European ancestry and AA participants of the age group 18 to 30 years, recruited from four centers. Details of the CARDIA study design have been previously published (Friedman et al. 1988). The CARDIA Women’s Study performed standardized study ultrasounds on women from CARDIA to detect the presence or absence of fibroids, as previously described (Wellons et al. 2008).
Stage 2 data were provided by the direct-to-consumer genetic testing company 23andMe, which used a survey for determining case or control status. Specifically, cases were defined as females responding positively to either of the following questions: “Has a doctor ever told you that you have uterine fibroids?”, or “Have you ever been diagnosed with uterine fibroids?”. Controls were defined as females responding with a ‘no’ to either of the questions above. Age at enrollment with 23andMe was also collected (mean ages of 56 in cases and 47 in controls).
In addition, in Stage 2, GWAS samples were provided by the Black Women’s Health Study (BWHS), a U.S. prospective cohort study of premenopausal women (aged 22–50 years) in which uterine fibroid diagnoses were ascertained by self-report, with medical record validation among a subset of cases (Wise et al. 2004, 2005b). BWHS participants reported whether they had been diagnosed with fibroids on the follow-up questionnaires every 2 years (1997–2009). The women reported the calendar year of their first diagnosis, and whether the diagnosis was confirmed by ultrasound or surgery. Controls were premenopausal women who reported no diagnosis of fibroids through 2009. Among those who released their medical records, 96% of self-reported fibroid cases were confirmed (Wise et al. 2005b). The study workflow is diagrammed in Fig. 1.
Genotyping
BioVU samples were genotyped using both the Affymetrix BioBank array and the Axiom World array 2 (Affymetrix Inc., Santa Clara, CA, USA) to attain better coverage for African-derived variants. BioME and the Mt. Sinai eMERGE site used the Illumina 1 M (Illumina Inc., San Diego, CA, USA) platform, with an exome chip backbone included for BioME samples. CARDIA-WS was genotyped on the Affymetrix 6.0 array (Affymetrix Inc., Santa Clara, CA, USA) as part of the Candidate-gene Association Resource which has been previously described in detail (Lettre et al. 2011).
The Stage 2 23andMe samples (1744 cases and 2906 controls) were genotyped on a custom GWAS panel across four versions. Stage 2 participants from BWHS (382 cases and 392 controls) were genotyped on the Illumina Infinium Expanded Multi-Ethnic Genotyping Array (MEGA; Illumina, Inc., San Diego, CA, USA) at Vanderbilt University VANTAGE Core Genotyping facility.
Quality control
Genotype quality control was performed within each study population, and a uniform protocol was implemented for all Stage 1 studies using PLINK (Purcell et al. 2007), including a 95% SNP and individual call rate threshold, removal of first-degree related individuals, sex checks, alignment of alleles to the genomic ‘+’ strand, and visualization of ancestry by principal components analysis using the Eigenstrat software (Price et al. 2006). No samples were excluded from analyses based on principal components.
For 23andMe in Stage 2, SNPs with Hardy–Weinberg equilibrium p value <10−20 were excluded, as were those with call rate <95%, or with large allele frequency discrepancies compared to European 1000 Genomes reference data. For BWHS in Stage 2, genotype quality control was also performed in PLINK and consisted of 95% SNP and 90% individual call rate thresholds, removal of first-degree related individuals, sex checks, alignment of alleles to the genomic ‘+’ strand, and visualization of ancestry by principal components analysis using the Eigenstrat software (Price et al. 2006). No samples were excluded from analyses based on principal components.
Statistical analysis
All demographic data were summarized and evaluated with Stata 14.0 (College Station, TX, USA).
The Stage 1 data were imputed to the 1000 Genomes phase 3 reference panel using SHAPEIT2 (Delaneau et al. 2012) for haplotype phasing and IMPUTE2 (Duan et al. 2013) for genotype imputation, with phasing and imputation performed separately for each data set. SNPTEST2 (Marchini et al. 2007) was used to perform single-variant association analyses for each cohort independently. Analyses were adjusted for the first five principal components in each sample to account for population substructure (Supplemental Fig. 1). Additional analysis adjusting for age was also performed, as was an analysis to avoid misclassification errors by restricting controls to peri-menopausal women (age ≥45) to remove women who were at increased risk of developing fibroids.
The Stage 2 data from 23andMe were imputed to the March 2012 “v3” release of 1000 Genomes reference haplotypes. Phasing and imputation were performed for each genotyping platform version separately, using Beagle4 (version 3.3.1) to phase batches of 8000–9000 individuals across chromosomal segments of no more than 10,000 genotyped single nucleotide polymorphisms (SNPs), with overlaps of 200 SNPs, and a high-performance version of Minimac5 for imputation of each phased segment against all-ethnicity 1000 Genomes haplotypes, using five rounds and 200 states for parameter estimation. Logistic regression was modeled using an additive genetic model adjusting for age and five principal components, reporting likelihood ratio test p values for association with genotypes.
Stage 2 also utilized 776 samples from BWHS which were imputed to the 1000 Genomes phase 3 reference panel using SHAPEIT2 (Delaneau et al. 2012) for haplotype phasing and IMPUTE2 (Duan et al. 2013) for genotype imputation, with phasing and imputation performed separately for each data set. SNPTEST2 (Marchini et al. 2007) was used to perform single-variant association analyses. Association analysis was performed using logistic regression of the imputed genotype data. Analyses were adjusted for ten principal components.
All association results for Stage 1 and across Stages 1–2 were meta-analyzed using fixed effects inverse-variance weighted meta-analysis in METAL (Willer et al. 2010). The final meta-analysis of all variants including all stages consisted of 3399 cases and 4764 controls.
To further evaluate the genetic association results in the context of gene expression, we employed the novel method MetaXcan (Barbeira et al. 2016), an extension of the PrediXcan method (Gamazon et al. 2015). PrediXcan conducts a test of association between phenotypes and gene expression levels predicted by genetic variants in a library of tissues from the Genotype-Tissue Expression (GTEx) project (2015; Carithers et al. 2015; Mele et al. 2015). MetaXcan is a meta-analysis approach that conducts the PrediXcan test using genotype association summary statistics, rather than performing the tests in individual-level data. For the purposes of this study, we utilized covariance matrices built for nine relevant tissues from GTEx (i.e., uterus, ovary, breast, vagina, subcutaneous adipose, visceral omentum adipose, thyroid, whole blood, and transformed fibroblasts) to annotate SNP association signals as well as to provide information about likely tissue expression patterns and relevant biological information.
Results
We used our previously validated EHR algorithm to define 999 image-confirmed cases and 1233 image-confirmed controls for fibroids from BioVU and Mt. Sinai (including the Mt. Sinai eMERGE subset as well as other cases and controls from the remainder of the BioME resource) sites. We also included additional samples from the CARDIA Women’s Study, which incorporated a standardized research ultrasound, for a total of 1273 case and 1379 control samples for analysis in Stage 1 discovery analyses (Fig. 1). Descriptive characteristics of each study are presented in Table 1. Briefly, cases and controls were of similar ages within each individual study in Stage 1 (no more than 4 year difference in means), though the Mt. Sinai sample was substantially older on average than the other three studies. However, comparing ages across studies is challenging due to the differing study designs (case–control vs. cohort) and incident periods.
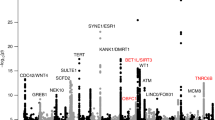
All data sets were genotyped on study-specific genome-wide genotyping arrays, imputed to the 1000 Genomes reference panel, and analyzed for association with fibroids, adjusting for principal components of ancestry (Supplemental Table 1). Meta-analysis of both Stages (3399 cases and 4764 controls) revealed a single genome-wide significant locus near CYTH4 (cytohesin 4) on chromosome 22 (Table 2; bottom panel of Fig. 2). The lead SNP at this locus, rs739187 [p value = 7.82 × 10−9, OR [95% confidence interval (CI)] = 1.23 (1.16–1.30)] had low heterogeneity across cohorts.
Top panel presents (as a mirror of the association results) the association with predicted gene expression levels from relevant tissues from GTEx, also arranged by chromosome and position; bottom panel presents SNP association results from meta-analysis across both stages in an inverted Manhattan plot format, arranged by chromosome and position
To further evaluate the genetic association results in the context of gene expression, we employed the novel method MetaXcan which conducts a test of association between phenotypes and gene expression levels predicted by genetic variants in a library of tissues from the GTEx project (GTEx Consortium et al. 2015; Carithers et al. 2015; Mele et al. 2015). Analysis with MetaXcan to evaluate association with genetically predicted gene expression (GPGE) levels resulted in 1542 results across 9 GTEx tissues with p value <0.05, from a total of 44,577 comparisons (Table 3; upper panel of Fig. 2). Examination of the locus implicated by the most significant genetic result, CYTH4, revealed a significant association with reduced GPGE at that gene in thyroid tissue (p value = 5.86 × 10−8). A striking result from this analysis was association between zinc finger protein 391 (ZNF391) GPGE in eight of the nine tissues and uterine fibroid risk (smallest p value = 5.05 × 10−5 in breast tissue).
Discussion
In this first multi-stage GWAS of uterine fibroids among AA women, we found a genome-wide significant result with linkage disequilibrium support on chromosome 22 near the CYTH4 gene (Fig. 3). Not a great deal is known about this gene, although variants near this locus have been previously suggestively associated with methotrexate response in juvenile idiopathic arthritis (Cobb et al. 2014), and a primate-specific repeat in the promoter of CYTH4 has been linked to bipolar disorder (Rezazadeh et al. 2015). CYTH4 is most highly expressed in whole blood and spleen based on observations in GTEx and only expressed at low levels in all other measured tissues. CYTH4 expression has also been observed to be low in myometrial tissue postpartum (Kanamarlapudi et al. 2012). The top signal from the GPGE analysis also identified predicted reduced CYTH4 in thyroid tissue as associated with fibroid risk, supporting a potential biological role for this gene. Women with fibroids have previously been shown to often have concurrent thyroid conditions, including overt hypothyroidism (Ott et al. 2014), thyroid nodules (Kim et al. 2010; Spinos et al. 2007), and thyroid cancer (Braganza et al. 2014; Makaridze and Mardaleishvili 2011). In addition, expression of genes related to fibroids (HMGA2 and PLAG1) was found to be correlated between fibroid and thyroid tumors (Klemke et al. 2014), which may provide an explanation for the observations in this study.
We also observed suggestive evidence of association (p ≤ 5 × 10−7) at a variant between LY6D (lymphocyte antigen 6 family member D) and JPH1 (junctophilin 1). The lead SNP in this region, rs6472827, is located 25 kb upstream of the coding region for JPH1 on chromosome 8, and is a cis-eQTL in both sun exposed and non-sun exposed skin tissue from GTEx. However, the GPGE results implicated decreased LY6D expression in breast tissue as associated with fibroid risk in the region (p value = 1.52 × 10−4). Increased expression of this gene has been observed in a variety of cancer types, including breast, compared to normal non-cancerous counterpart tissues (Luo et al. 2016).
Evaluation of the genetic association results in the context of GPGE across a variety of tissues provided supportive evidence for the biological mechanism underlying the genetic data. Perhaps most striking among GPGE results was ZNF391, which encodes zinc finger protein 391, which was suggestive in eight of the nine tissues (and nominal in uterus, p value = 0.03). This gene has previously been implicated in rheumatoid arthritis (Orozco et al. 2011), and more interestingly, is expressed in several reproductive-related tissues in GTEx. The highest expression of ZNF391 is in testis and ovary, with uterine expression falling in the top third of tissues. Furthermore, a SNP in this gene (rs4713108) is suggestively associated with expression of ZNF391 in uterine tissue (p value = 1.1 × 10−7). This particular SNP was not associated with fibroid risk in our meta-analysis (p value = 0.08); however, there was a broad base of suggestive SNPs in the region with p values as low as 2.01 × 10−6 (Supplemental Fig. 2).
Another gene target identified through GPGE, ALDH2 (p value = 7.30 × 10−5), encodes a mitochondrial isoform of alcohol dehydrogenase, which is involved in alcohol metabolism. Five studies have shown a consistent relationship between increased alcohol consumption and fibroid risk (OR = 1.25–2.78) (D’Aloisio and Baird 2004; Marshall et al. 1997; Nagata et al. 2009; Templeman et al. 2009; Wise et al. 2004). Predicted decreased levels of AGT in thyroid tissue were also implicated as being associated with fibroid risk (p value = 2.49 × 10−5). AGT encodes angiotensinogen, which is involved in renal homeostasis, and given the observed link between hypertension and fibroids, may support a plausible biological connection between the two.
Previous GWAS for fibroids in Japanese subjects has implicated variants near the BET1L, TNRC6B, and OBFC1–SLK genes (Cha et al. 2011). We did not observe suggestive associations in our AA subjects at the prior GWAS-associated SNPs near BET1L and OBFC1-SLK which were reported by Cha et al. (Supplemental Figs. 3–8). The best result at these loci was near TNRC6B (rs59426214; p value = 2.42 × 10−5; Supplemental Fig. 3), although this variant was not in LD with the index SNP (rs12484776) detected by Cha et al. This lack of strong association is largely consistent with prior published studies that have attempted to replicate the associations within cohorts of African ancestry.
A potential consideration of this study was that the Stage 2 studies relied upon self-reported fibroids information, which is known to be subject to misclassification bias. However, the top result at CYTH4 was suggestive in the imaged Stage 1 analysis with a p value of 6.46 × 10−7, and effect sizes were consistent between Stages 1 and 2. Additional points to consider are that the control group was not restricted to women beyond menopause, when they would have a negligible likelihood of developing new fibroids. We did explore this as a secondary analysis in the studies from Stage 1, albeit with reduced power due to the smaller sample size (data not shown). Restricting the controls to women peri-menopausal and younger (age <45) did not change the overall associations, as many of the upper tier signals were of similar effect size (r 2 = 0.87) despite the lack of significance due to reduced power. In addition, adjustment for age in Stage 1 did not markedly alter the results seen in the primary analysis, with several of the top overall signals remaining suggestively significant (6/12 variants with p value <1e−4) and having consistent effect sizes (r 2 = 0.96). Further considerations are the small numbers and heterogeneity among tissues included in the GTEx models. Uterus in particular had a very small number of tissue samples (N = 70), and is known to be a heterogeneous mix of specific cell/tissue types.
In conclusion, in this first multi-stage GWAS of uterine fibroids among AA women, we identified a novel genome-wide significant result in CYTH4 with biological support from the significant GPGE showing reduced CYTH4 expression in thyroid tissue. Further studies are needed to confirm the SNP and target loci and elucidate the biological role of this gene and others identified through GWAS and GPGE in relation with fibroid development (Edwards et al. 2013a). Overall, we have identified novel genetic signals, highlighting the importance of this work for understanding fibroids in AA women.
Change history
04 October 2017
The article “A multi-stage genome-wide association study of uterine fibroids in African Americans”, written by Jacklyn N. Hellwege, was originally published Online First without open access. After publication in volume 136, issue 10, page 1363–1373 the author decided to opt for Open Choice and to make the article an open access publication. Therefore, the copyright of the article has been changed to © The Author(s) 2017 and the article is forthwith distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
References
Aissani B, Zhang K, Wiener H (2015a) Evaluation of GWAS candidate susceptibility loci for uterine leiomyoma in the multi-ethnic NIEHS uterine fibroid study. Front Genet 6:241. doi:10.3389/fgene.2015.00241
Aissani B, Zhang K, Wiener H (2015b) Follow-up to genome-wide linkage and admixture mapping studies implicates components of the extracellular matrix in susceptibility to and size of uterine fibroids. Fertil Steril 103(528–34):e13. doi:10.1016/j.fertnstert.2014.10.025
Baird DD, Dunson DB (2003) Why is parity protective for uterine fibroids? Epidemiology 14:247–250. doi:10.1097/01.ede.0000054360.61254.27
Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM (2003) High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol 188:100–107
Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM (2007) Association of physical activity with development of uterine leiomyoma. Am J Epidemiol 165:157–163. doi:10.1093/aje/kwj363
Barbeira A, Dickinson SP, Torres JM, Torstenson ES, Zheng J, Wheeler HE, Shah KP, Edwards T, Consortium G, Nicolae D, Cox NJ, Im HK (2016) Integrating tissue specific mechanisms into GWAS summary results. bioRxiv. doi:10.1101/045260
Borah BJ, Nicholson WK, Bradley L, Stewart EA (2013) The impact of uterine leiomyomas: a national survey of affected women. Am J Obstet Gynecol 209:319.e1–319.e20. doi:10.1016/j.ajog.2013.07.017
Braganza MZ, de Gonzalez AB, Schonfeld SJ, Wentzensen N, Brenner AV, Kitahara CM (2014) Benign breast and gynecologic conditions, reproductive and hormonal factors, and risk of thyroid cancer. Cancer Prev Res (Phila) 7:418–425. doi:10.1158/1940-6207.capr-13-0367
Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, Guan P, Korzeniewski GE, Lockhart NC, Rabiner CA, Rao AK, Robinson KL, Roche NV, Sawyer SJ, Segre AV, Shive CE, Smith AM, Sobin LH, Undale AH, Valentino KM, Vaught J, Young TR, Moore HM (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank 13:311–319. doi:10.1089/bio.2015.0032
Cha PC, Takahashi A, Hosono N, Low SK, Kamatani N, Kubo M, Nakamura Y (2011) A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat Genet 43:447–450. doi:10.1038/ng.805
Cobb J, Cule E, Moncrieffe H, Hinks A, Ursu S, Patrick F, Kassoumeri L, Flynn E, Bulatovic M, Wulffraat N, van Zelst B, de Jonge R, Bohm M, Dolezalova P, Hirani S, Newman S, Whitworth P, Southwood TR, De Iorio M, Wedderburn LR, Thomson W (2014) Genome-wide data reveal novel genes for methotrexate response in a large cohort of juvenile idiopathic arthritis cases. Pharmacogenomics J 14:356–364. doi:10.1038/tpj.2014.3
D’Aloisio AA, Baird D (2004) Variation in the association of alcohol intake with uterine fibroids by race and tumor size. In: American College of Epidemiology, vol 14. Annals of epidemiology, Boston, p 622
Delaneau O, Marchini J, Zagury JF (2012) A linear complexity phasing method for thousands of genomes. Nat Methods 9:179–181. doi:10.1038/nmeth.1785
Duan Q, Liu EY, Auer PL, Zhang G, Lange EM, Jun G, Bizon C, Jiao S, Buyske S, Franceschini N, Carlson CS, Hsu L, Reiner AP, Peters U, Haessler J, Curtis K, Wassel CL, Robinson JG, Martin LW, Haiman CA, Le Marchand L, Matise TC, Hindorff LA, Crawford DC, Assimes TL, Kang HM, Heiss G, Jackson RD, Kooperberg C, Wilson JG, Abecasis GR, North KE, Nickerson DA, Lange LA, Li Y (2013) Imputation of coding variants in African Americans: better performance using data from the exome sequencing project. Bioinformatics 29:2744–2749. doi:10.1093/bioinformatics/btt477
Edwards SL, Beesley J, French JD, Dunning AM (2013a) Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93:779–797. doi:10.1016/j.ajhg.2013.10.012
Edwards TL, Michels KA, Hartmann KE, Velez Edwards DR (2013b) BET1L and TNRC6B associate with uterine fibroid risk among European Americans. Hum Genet 132:943–953. doi:10.1007/s00439-013-1306-3
Eggert SL, Huyck KL, Somasundaram P, Kavalla R, Stewart EA, Lu AT, Painter JN, Montgomery GW, Medland SE, Nyholt DR, Treloar SA, Zondervan KT, Heath AC, Madden PA, Rose L, Buring JE, Ridker PM, Chasman DI, Martin NG, Cantor RM, Morton CC (2012) Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. Am J Hum Genet 91:621–628. doi:10.1016/j.ajhg.2012.08.009
Faerstein E, Szklo M, Rosenshein N (2001) Risk factors for uterine leiomyoma: a practice-based case-control study. I. African-American heritage, reproductive history, body size, and smoking. Am J Epidemiol 153:1–10
Feingold-Link L, Edwards TL, Jones S, Hartmann KE, Velez Edwards DR (2014) Enhancing uterine fibroid research through utilization of biorepositories linked to electronic medical record data. J Womens Health (Larchmt) 23:1027–1032. doi:10.1089/jwh.2014.4978
Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, Liu K, Savage PJ (1988) CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 41:1105–1116
Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47:1091–1098. doi:10.1038/ng.3367
Genotype-Tissue Expression (GTEx) (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. doi:10.1126/science.1262110
Kanamarlapudi V, Owens SE, Lartey J, Lopez Bernal A (2012) ADP-ribosylation factor 6 expression and activation are reduced in myometrium in complicated pregnancies. PLoS One 7:e37954. doi:10.1371/journal.pone.0037954
Kim MH, Park YR, Lim DJ, Yoon KH, Kang MI, Cha BY, Lee KW, Son HY (2010) The relationship between thyroid nodules and uterine fibroids. Endocr J 57:615–621
Klemke M, Muller MH, Wosniok W, Markowski DN, Nimzyk R, Helmke BM, Bullerdiek J (2014) Correlated expression of HMGA2 and PLAG1 in thyroid tumors, uterine leiomyomas and experimental models. PLoS One 9:e88126. doi:10.1371/journal.pone.0088126
Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, Farlow DN, Folsom AR, Fornage M, Forrester T, Fox E, Haiman CA, Hartiala J, Harris TB, Hazen SL, Heckbert SR, Henderson BE, Hirschhorn JN, Keating BJ, Kritchevsky SB, Larkin E, Li M, Rudock ME, McKenzie CA, Meigs JB, Meng YA, Mosley TH, Newman AB, Newton-Cheh CH, Paltoo DN, Papanicolaou GJ, Patterson N, Post WS, Psaty BM, Qasim AN, Qu L, Rader DJ, Redline S, Reilly MP, Reiner AP, Rich SS, Rotter JI, Liu Y, Shrader P, Siscovick DS, Tang WH, Taylor HA, Tracy RP, Vasan RS, Waters KM, Wilks R, Wilson JG, Fabsitz RR, Gabriel SB, Kathiresan S, Boerwinkle E (2011) Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet 7:e1001300. doi:10.1371/journal.pgen.1001300
Luo L, McGarvey P, Madhavan S, Kumar R, Gusev Y, Upadhyay G (2016) Distinct lymphocyte antigens 6 (Ly6) family members Ly6D, Ly6E, Ly6K and Ly6H drive tumorigenesis and clinical outcome. Oncotarget 7:11165–11193. doi:10.18632/oncotarget.7163
Luoto R, Kaprio J, Rutanen EM, Taipale P, Perola M, Koskenvuo M (2000) Heritability and risk factors of uterine fibroids–the Finnish Twin Cohort study. Maturitas 37:15–26
Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39(7):906–913. doi:10.1038/ng2088
Makaridze T, Mardaleishvili K (2011) Combined development of thyroid gland and reproductive system benign diseases. Georgian Med News (199):20–29
Marshall LM, Spiegelman D, Barbieri RL, Goldman MB, Manson JE, Colditz GA, Willett WC, Hunter DJ (1997) Variation in the incidence of uterine leiomyoma among premenopausal women by age and race. Obstet Gynecol 90:967–973
Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segre AV, Djebali S, Niarchou A, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigo R (2015) Human genomics. The human transcriptome across tissues and individuals. Science 348:660–665. doi:10.1126/science.aaa0355
Moore AB, Flake GP, Swartz CD, Heartwell G, Cousins D, Haseman JK, Kissling GE, Sidawy MK, Dixon D (2008) Association of race, age and body mass index with gross pathology of uterine fibroids. J Reprod Med 53:90–96
Myers SL, Baird DD, Olshan AF, Herring AH, Schroeder JC, Nylander-French LA, Hartmann KE (2012) Self-report versus ultrasound measurement of uterine fibroid status. J Womens Health (Larchmt) 21:285–293. doi:10.1089/jwh.2011.3008
Nagata C, Nakamura K, Oba S, Hayashi M, Takeda N, Yasuda K (2009) Association of intakes of fat, dietary fibre, soya isoflavones and alcohol with uterine fibroids in Japanese women. Br J Nutr 101:1427–1431
Orozco G, Barton A, Eyre S, Ding B, Worthington J, Ke X, Thomson W (2011) HLA-DPB1-COL11A2 and three additional xMHC loci are independently associated with RA in a UK cohort. Genes Immun 12:169–175. doi:10.1038/gene.2010.57
Ott J, Kurz C, Braun R, Promberger R, Seemann R, Vytiska-Binstorfer E, Walch K (2014) Overt hypothyroidism is associated with the presence of uterine leiomyoma: a retrospective analysis. Eur J Obstet Gynecol Reprod Biol 177:19–22. doi:10.1016/j.ejogrb.2014.03.003
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. doi:10.1038/ng1847
Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR (2010) Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 3:42–48. doi:10.1111/j.1752-8062.2010.00175.x
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi:10.1086/519795
Rezazadeh M, Gharesouran J, Mirabzadeh A, Khorram Khorshid HR, Biglarian A, Ohadi M (2015) A primate-specific functional GTTT-repeat in the core promoter of CYTH4 is linked to bipolar disorder in human. Prog Neuropsychopharmacol Biol Psychiatry 56:161–167. doi:10.1016/j.pnpbp.2014.09.001
Spinos N, Terzis G, Crysanthopoulou A, Adonakis G, Markou KB, Vervita V, Koukouras D, Tsapanos V, Decavalas G, Kourounis G, Georgopoulos NA (2007) Increased frequency of thyroid nodules and breast fibroadenomas in women with uterine fibroids. Thyroid 17:1257–1259. doi:10.1089/thy.2006.0330
Templeman C, Marshall SF, Clarke CA, Henderson KD, Largent J, Neuhausen S, Reynolds P, Ursin G, Bernstein L (2009) Risk factors for surgically removed fibroids in a large cohort of teachers. Fertil Steril 92:1436–1446. doi:10.1016/j.fertnstert.2008.08.074
Terry KL, De Vivo I, Hankinson SE, Spiegelman D, Wise LA, Missmer SA (2007) Anthropometric characteristics and risk of uterine leiomyoma. Epidemiology 18:758–763. doi:10.1097/EDE.0b013e3181567eed
Vollenhoven B (1998) Introduction: the epidemiology of uterine leiomyomas. Baillieres Clin Obstet Gynaecol 12:169–176
Wellons MF, Lewis CE, Schwartz SM, Gunderson EP, Schreiner PJ, Sternfeld B, Richman J, Sites CK, Siscovick DS (2008) Racial differences in self-reported infertility and risk factors for infertility in a cohort of black and white women: the CARDIA Women’s Study. Fertil Steril 90:1640–1648. doi:10.1016/j.fertnstert.2007.09.056
Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191. doi:10.1093/bioinformatics/btq340
Wise LA, Palmer JR, Harlow BL, Spiegelman D, Stewart EA, Adams-Campbell LL, Rosenberg L (2004) Risk of uterine leiomyomata in relation to tobacco, alcohol and caffeine consumption in the Black Women’s Health Study. Hum Reprod 19:1746–1754. doi:10.1093/humrep/deh309
Wise LA, Palmer JR, Spiegelman D, Harlow BL, Stewart EA, Adams-Campbell LL, Rosenberg L (2005a) Influence of body size and body fat distribution on risk of uterine leiomyomata in U.S. black women. Epidemiology 16:346–354
Wise LA, Palmer JR, Stewart EA, Rosenberg L (2005b) Age-specific incidence rates for self-reported uterine leiomyomata in the Black Women’s Health Study. Obstet Gynecol 105:563–568. doi:10.1097/01.AOG.0000154161.03418.e3
Wise LA, Ruiz-Narvaez EA, Palmer JR, Cozier YC, Tandon A, Patterson N, Radin RG, Rosenberg L, Reich D (2012) African ancestry and genetic risk for uterine leiomyomata. Am J Epidemiol 176:1159–1168. doi:10.1093/aje/kws276
Zhang K, Wiener H, Aissani B (2015) Admixture mapping of genetic variants for uterine fibroids. J Hum Genet. doi:10.1038/jhg.2015.60
Acknowledgements
This work was supported by the National Institutes of Health Grants R01HD074711 (DRVE), R03HD078567 (DRVE), U01HG08672-01 (DMR), and R01HD060530 (CCM), as well as the Vanderbilt Molecular and Genetic Epidemiology of Cancer (MAGEC) training program, funded by R25CA160056 (PI: X.-O. Shu). One of the data sets used for the analyses described was obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding, the 1S10RR025141-01 instrumentation award, and by the Vanderbilt CTSA Grant UL1TR000445 from NCATS/NIH. The BioMe Biobank program is supported by The Andrea and Charles Bronfman Philanthropies. The Electronic Medical Records and Genomics Network was initiated and funded by the National Human Genome Research Institute through by eMERGE Network Grant U01HG006380 to Mt. Sinai. The Coronary Artery Risk Development in Young Adults Study (CARDIA) is supported by contracts HHSN268201300025C, HHSN268201300026C, HHSN268201300027C, HHSN268201300028C, HHSN268201300029C, and HHSN268200900041C from the National Heart, Lung, and Blood Institute (NHLBI), the Intramural Research Program of the National Institute on Aging (NIA), and an intra-agency agreement between NIA and NHLBI (AG0005). Genotyping was funded as part of the NHLBI Candidate-gene Association Resource (N01-HC-65226). Uterine fibroid-related work from the Black Women’s Health Study (BWHS) was supported by Grants R01CA58420 (LR), UM1CA164974 (LR), and R01CA098663 (JP) from the National Cancer Institute, and Grant R01HD057966 (LAW). The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health. We thank the research participants and staff from all studies, as well as the employees of 23andMe, for making this work possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DAH is an employee of and owns stock options in 23andMe, Inc. EAS declares the following financial competing interests: consultancy related to uterine fibroids: AbbVie, Allergan, Astellas, Bayer, and Gynesonics, Viteava. Patent applications (pending or actual), including individual applications or those belonging to the institution to which the authors, are affiliated and from which the authors may benefit. US 6440445 (With Nowak, RA, issued 2002, no commercial activity). Research Grants: NIH. R01HD060503, PI. P50 HS023418, Site PI for 3 component parts. R01HD074711, Consultant. K12HD065987, Board Member for Mayo Clinic Site. Other: Focused Ultrasound Surgery Foundation Borah(PI) 11/15/11-11/14/12 Costs of Uterine Fibroid Treatments Including Focused Ultrasound Surgery. Insightec Inc, PI Patient care costs in conjunction with R01HD60530. In addition, the following non-financial competing interests: relationship (paid or unpaid) with organizations and funding bodies including nongovernmental organizations, research institutions, or charities. Registry Steering Committee, REgistry for Leiomyoma International Efficacy of Focused Ultrasound (RELIEF). Writing or consulting for an educational company. Honoraria from: UpToDate, Massachusetts Medical Society.
Additional information
An erratum to this article is available at https://doi.org/10.1007/s00439-017-1846-z.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hellwege, J.N., Jeff, J.M., Wise, L.A. et al. A multi-stage genome-wide association study of uterine fibroids in African Americans. Hum Genet 136, 1363–1373 (2017). https://doi.org/10.1007/s00439-017-1836-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1836-1