Abstract
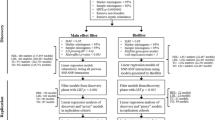
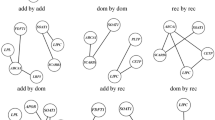
Genetic loci explain only 25–30 % of the heritability observed in plasma lipid traits. Epistasis, or gene–gene interactions may contribute to a portion of this missing heritability. Using the genetic data from five NHLBI cohorts of 24,837 individuals, we combined the use of the quantitative multifactor dimensionality reduction (QMDR) algorithm with two SNP-filtering methods to exhaustively search for SNP–SNP interactions that are associated with HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), total cholesterol (TC) and triglycerides (TG). SNPs were filtered either on the strength of their independent effects (main effect filter) or the prior knowledge supporting a given interaction (Biofilter). After the main effect filter, QMDR identified 20 SNP–SNP models associated with HDL-C, 6 associated with LDL-C, 3 associated with TC, and 10 associated with TG (permutation P value <0.05). With the use of Biofilter, we identified 2 SNP–SNP models associated with HDL-C, 3 associated with LDL-C, 1 associated with TC and 8 associated with TG (permutation P value <0.05). In an independent dataset of 7502 individuals from the eMERGE network, we replicated 14 of the interactions identified after main effect filtering: 11 for HDL-C, 1 for LDL-C and 2 for TG. We also replicated 23 of the interactions found to be associated with TG after applying Biofilter. Prior knowledge supports the possible role of these interactions in the genetic etiology of lipid traits. This study also presents a computationally efficient pipeline for analyzing data from large genotyping arrays and detecting SNP–SNP interactions that are not primarily driven by strong main effects.



Similar content being viewed by others
References
Akram ON, Bernier A, Petrides F et al (2010) Beyond LDL cholesterol, a new role for PCSK9. Arterioscler Thromb Vasc Biol 30:1279–1281. doi:10.1161/ATVBAHA.110.209007
Ansari KI, Kasiri S, Hussain I et al (2012) MLL histone methylases regulate expression of HDLR-SR-B1 in presence of estrogen and control plasma cholesterol in vivo. Mol Endocrinol 27:92–105. doi:10.1210/me.2012-1147
Arsenault BJ, Boekholdt SM, Kastelein JJP (2011) Lipid parameters for measuring risk of cardiovascular disease. Nat Rev Cardiol 8:197–206
Asselbergs FW, Guo Y, Van Iperen EPA et al (2012) Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet 91:823–838. doi:10.1016/j.ajhg.2012.08.032
Aung LH, Yin RX, Wu JZ et al (2014) Association between the MLX interacting protein-like, BUD13 homolog and zinc finger protein 259 gene polymorphisms and serum lipid levels. Sci Rep 4:5565. doi:10.1038/srep05565
Barter PJ, Brewer HB, Chapman MJ et al (2003) Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler Thromb Vasc Biol 23:160–167. doi:10.1161/01.ATV.0000054658.91146.64
Benn M, Nordestgaard BG, Jensen JS et al (2005) Polymorphism in APOB associated with increased low-density lipoprotein levels in both genders in the general population. J Clin Endocrinol Metab 90:5797–5803. doi:10.1210/jc.2005-0974
Bild DE, Bluemke DA, Burke GL et al (2002) Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol 156:871–881
Brooks MA, Dziembowski A, Quevillon-Cheruel S et al (2009) Structure of the yeast Pml1 splicing factor and its integration into the RES complex. Nucleic Acids Res 37:129–143. doi:10.1093/nar/gkn894
Brown ML, Inazu A, Hesler CB et al (1989) Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature 342:448–451. doi:10.1038/342448a0
Bush WS, Dudek SM, Ritchie MD (2009) Biofilter: A knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pacific Symp Biocomput 368–379
Bush WS, McCauley JL, DeJager PL et al (2011) A knowledge-driven interaction analysis reveals potential neurodegenerative mechanism of multiple sclerosis susceptibility. Genes Immun 12:335–340. doi:10.1038/gene.2011.3
Calle ML, Urrea V, Malats N, Van Steen K (2010) mbmdr: an R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics 26:2198–2199. doi:10.1093/bioinformatics/btq352
Cao A, Wu M, Li H, Liu J (2011) Janus kinase activation by cytokine oncostatin M decreases PCSK9 expression in liver cells. J Lipid Res 52:518–530. doi:10.1194/jlr.M010603
Dawber TR, Meadors GF, Moore FE (1951) Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health 41:279–281
Deaton C, Froelicher ES, Wu LH et al (2011) The global burden of cardiovascular disease. Eur J Cardiovasc Nurs 10:S5–S13. doi:10.1016/S1474-5151(11)00111-3
Eichler EE, Flint J, Gibson G et al (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11:446–450. doi:10.1038/nrg2809
Fried LP, Borhani NO, Enright P et al (1991) The cardiovascular health study: design and rationale. Ann Epidemiol 1:263–276
Friedewald WT, Levy RI, Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18:499–502
Friedman GD, Cutter GR, Donahue RP et al (1988) CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 41:1105–1116
Gottesman O, Kuivaniemi H, Tromp G et al (2013) The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet Med 15:761–771. doi:10.1038/gim.2013.72
Grady BJ, Torstenson ES, McLaren PJ et al (2011) Use of biological knowledge to inform the analysis of gene-gene interactions involved in modulating virologic failure with efavirenz-containing treatment regimens in ART-naive ACTG clinical trials participants. Pac Symp Biocomput 253–264
Gui J, Moore JH, Williams SM et al (2013) A simple and computationally efficient approach to multifactor dimensionality reduction analysis of gene-gene interactions for quantitative traits. PLoS One 8:e66545. doi:10.1371/journal.pone.0066545
Gundlach S, Kässens JC, Wienbrandt L (2016) Genome-wide association interaction studies with MB-MDR and maxT multiple testing correction on FPGAs. Procedia Comput Sci 80:639–649. doi:10.1016/j.procs.2016.05.354
Hall MA, Verma SS, Wallace J et al (2015) Biology-driven gene-gene interaction analysis of age-related cataract in the eMERGE network. Genet Epidemiol 39:376–384. doi:10.1002/gepi.21902
Heller DA, de Faire U, Pedersen NL et al (1993) Genetic and environmental influences on serum lipid levels in twins. N Engl J Med 328:1150–1156. doi:10.1056/NEJM199304223281603
Hill C, Gerardo D, James F et al (1989) The Atherosclerosis risk in communities (ARIC) study: design and objectives. Am J Epidemiol 129:687–702
Hooper AJ, van Bockxmeer FM, Burnett JR (2005) Monogenic hypocholesterolaemic lipid disorders and apolipoprotein B metabolism. Crit Rev Clin Lab Sci 42:515–545. doi:10.1080/10408360500295113
Horton JD, Goldstein JL, Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131. doi:10.1172/JCI15593
Ide T, Shimano H, Yahagi N et al (2004) SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol 6:351–357
Johnson AD, Handsaker RE, Pulit SL et al (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24:2938–2939. doi:10.1093/bioinformatics/btn564
Kathiresan S, Melander O, Guiducci C et al (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197
Kathiresan S, Willer CJ, Peloso GM et al (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41:56–65. doi:10.1038/ng.291
Keating BJ, Tischfield S, Murray SS et al (2008) Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One 3:e3583. doi:10.1371/journal.pone.0003583
Klos K, Shimmin L, Ballantyne C et al (2008) APOE/C1/C4/C2 hepatic control region polymorphism influences plasma apoE and LDL cholesterol levels. Hum Mol Genet 17:2039–2046. doi:10.1093/hmg/ddn101
Kuivenhoven JA, de Knijff P, Boer JMA et al (1997) Heterogeneity at the CETP gene locus: influence on plasma CETP concentrations and HDL cholesterol levels. Arterioscler Thromb Vasc Biol 17:560–568. doi:10.1161/01.ATV.17.3.560
Ma L, Yang J, Runesha HB et al (2010) Genome-wide association analysis of total cholesterol and high-density lipoprotein cholesterol levels using the Framingham heart study data. BMC Med Genet 11:55. doi:10.1186/1471-2350-11-55
Manolio TA, Collins FS, Cox NJ et al (2009) Finding the missing heritability of complex diseases. Nature 461:747–753. doi:10.1038/nature08494
McCarty CA, Chisholm RL, Chute CG et al (2011) The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 4:13. doi:10.1186/1755-8794-4-13
Middelberg RPS, Ferreira MAR, Henders AK et al (2011) Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet 12:123. doi:10.1186/1471-2350-12-123
Moore JH, Asselbergs FW, Williams SM (2010) Bioinformatics challenges for genome-wide association studies. Bioinformatics 26:445–455. doi:10.1093/bioinformatics/btp713
Nelson ER, Chang C, McDonnell DP (2014) Cholesterol and breast cancer pathophysiology. Trends Endocrinol Metab 25:649–655. doi:10.1016/j.tem.2014.10.001
Olsson AH, Volkov P, Bacos K et al (2014) Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet 10:e1004735. doi:10.1371/journal.pgen.1004735
Paplomata E, O’Regan R (2014) The PI3 K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol 6:154–166. doi:10.1177/1758834014530023
Pendergrass SA, Frase A, Wallace J, et al (2013) Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. BioData Min 6:25. doi:10.1186/1756-0381-6-25
Price AL, Patterson NJ, Plenge RM et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. doi:10.1038/ng1847
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi:10.1086/519795
Rasmussen-Torvik LJ, Pacheco JA, Wilke RA et al (2012) High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin Transl Sci 5:394–399. doi:10.1111/j.1752-8062.2012.00446.x
Reymer PW, Gagne E, Groenemeyer BE et al (1995) A lipoprotein lipase mutation (Asn291Ser) is associated with reduced HDL cholesterol levels in premature atherosclerosis. Nat Genet 10:28–34. doi:10.1038/ng0595-28
Ritchie MD (2011) Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann Hum Genet 75:172–182. doi:10.1111/j.1469-1809.2010.00630.x.Using
Ritchie MD, Hahn LW, Roodi N et al (2001) Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 69:138–147. doi:10.1086/321276
Soto-Ramírez N, Arshad SH, Holloway JW et al (2013) The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin Epigenetics 5:1. doi:10.1186/1868-7083-5-1
Sun X, Lu Q, Mukheerjee S et al (2014) Analysis pipeline for the epistasis search—statistical versus biological filtering. Front Genet 5:106. doi:10.3389/fgene.2014.00106
Talmud PJ, Drenos F, Shah S et al (2009) Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet 85:628–642. doi:10.1016/j.ajhg.2009.10.014
Teslovich TM, Musunuru K, Smith AV et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713. doi:10.1038/nature09270
Turner SD, Berg RL, Linneman JG et al (2011) Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PLoS One 6:e19586. doi:10.1371/journal.pone.0019586
Verma SS, de Andrade M, Tromp G et al (2014) Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet 5:1–15. doi:10.3389/fgene.2014.00370
Voight BF, Peloso GM, Orho-Melander M et al (2012) Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380:572–580. doi:10.1016/S0140-6736(12)60312-2
Wan X, Yang C, Yang Q et al (2010) BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am J Hum Genet 87:325–340. doi:10.1016/j.ajhg.2010.07.021
Waterworth DM, Ricketts SL, Song K et al (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 30:2264–2276. doi:10.1161/ATVBAHA.109.201020
Weiss LA, Pan L, Abney M, Ober C (2006) The sex-specific genetic architecture of quantitative traits in humans. Nat Genet 38:218–222. doi:10.1038/ng1726
Wittekoek ME, Pimstone SN, Reymer PWA et al (1998) A common mutation in the lipoprotein lipase gene (N291S) alters the lipoprotein phenotype and risk for cardiovascular disease in patients with familial hypercholesterolemia. Circulation 97:729–735. doi:10.1161/01.CIR.97.8.729
Wong AK, Park CY, Greene CS et al (2012) IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res 40:W484–W490. doi:10.1093/nar/gks458
World Health Organization (2014) Mendis, Shanthi. Global status report on noncommunicable diseases 2014, p 9
Xu M, Li X, Wang J-G et al (2005) Glucose and lipid metabolism in relation to novel polymorphisms in the 5′-AMP-activated protein kinase gamma2 gene in Chinese. Mol Genet Metab 86:372–378. doi:10.1016/j.ymgme.2005.06.012
Yao J, Yan M, Guan Z et al (2009) Aurora-A down-regulates IkappaBalpha via Akt activation and interacts with insulin-like growth factor-1 induced phosphatidylinositol 3-kinase pathway for cancer cell survival. Mol Cancer 8:95. doi:10.1186/1476-4598-8-95
Acknowledgments
CARe acknowledges the support of the National Heart, Lung and Blood Institute and the contributions of the research institutions, study investigators, field staff, and study participants in creating this resource for biomedical research (NHLBI contract number HHSN268200960009C). The IBC array data (also known as ‘Cardiochip’ or ‘CVDSNP55v1_A’) from the National Heart, Lung and Blood Institute’s (NHLBI) Candidate Gene Association Resource (CARe) was downloaded with appropriate permissions from the database of Genotypes and Phenotypes (dbGaP) (http://www.ncbi.nlm.gov/gap). The imputed genotype data for eMERGE-I and eMERGE-II can be downloaded from the database of Genotypes and Phenotypes (dbGaP) (http://www.ncbi.nlm.gov/gap).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding statement
This work was supported by National Institutes of Health grants: NLM R01 grants (LM0l0098, LM011360, LM009012), GMS P20 grants (GM103506, GM103534 and GM104416), and F31 HG008588. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
eMERGE Network (Phase II—Year 1) Acknowledgement
The eMERGE Network was initiated and funded by the National Human Genome Research Institute (NHGRI) through the following grants: U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) serving as Genotyping Centers.
eMERGE Network (Phase I) Acknowledgement
The eMERGE Network was initiated and funded by the National Human Genome Research Institute (NHGRI), in conjunction with additional funding from the National Institute of General Medical Sciences (NIGMS) through the following grants: U01-HG-004610 (Group Health Cooperative/University of Washington); U01-HG-004608 (Marshfield Clinic Research Foundation and Vanderbilt University Medical Center); U01-HG-04599 (Mayo Clinic); U01HG004609 (Northwestern University); U01-HG-04603 (Vanderbilt University Medical Center, also serving as the Administrative Coordinating Center); U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) serving as Genotyping Centers.
Conflict of interests
The authors declare that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
De, R., Verma, S.S., Holzinger, E. et al. Identifying gene–gene interactions that are highly associated with four quantitative lipid traits across multiple cohorts. Hum Genet 136, 165–178 (2017). https://doi.org/10.1007/s00439-016-1738-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1738-7