Abstract
Myopia is a complex genetic disorder and a common cause of visual impairment among working age adults. Genome-wide association studies have identified susceptibility loci on chromosomes 15q14 and 15q25 in Caucasian populations of European ancestry. Here, we present a confirmation and meta-analysis study in which we assessed whether these two loci are also associated with myopia in other populations. The study population comprised 31 cohorts from the Consortium of Refractive Error and Myopia (CREAM) representing 4 different continents with 55,177 individuals; 42,845 Caucasians and 12,332 Asians. We performed a meta-analysis of 14 single nucleotide polymorphisms (SNPs) on 15q14 and 5 SNPs on 15q25 using linear regression analysis with spherical equivalent as a quantitative outcome, adjusted for age and sex. We calculated the odds ratio (OR) of myopia versus hyperopia for carriers of the top-SNP alleles using a fixed effects meta-analysis. At locus 15q14, all SNPs were significantly replicated, with the lowest P value 3.87 × 10−12 for SNP rs634990 in Caucasians, and 9.65 × 10−4 for rs8032019 in Asians. The overall meta-analysis provided P value 9.20 × 10−23 for the top SNP rs634990. The risk of myopia versus hyperopia was OR 1.88 (95 % CI 1.64, 2.16, P < 0.001) for homozygous carriers of the risk allele at the top SNP rs634990, and OR 1.33 (95 % CI 1.19, 1.49, P < 0.001) for heterozygous carriers. SNPs at locus 15q25 did not replicate significantly (P value 5.81 × 10−2 for top SNP rs939661). We conclude that common variants at chromosome 15q14 influence susceptibility for myopia in Caucasian and Asian populations world-wide.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Refractive errors are common optical defects of the visual system. An important refractive error is myopia (nearsightedness), which occurs when the eye elongates beyond the focal plane. The prevalence of myopia is high, affecting about one-third of the world’s population, and reaching over 70 % in certain Asian ethnic groups (He et al. 2004; Kempen et al. 2004; Lin et al. 2004; Vitale et al. 2008; Wu et al. 2001). High degrees of myopia are associated with pathologic ocular changes, such as myopic macular degeneration, retinal detachment, and glaucoma (Curtin and Karlin 1971; McBrien and Gentle 2003; Saw 2006; Saw et al. 2005; Tano 2002). Due to the limited treatment options, myopia is a common cause of visual impairment (Tano 2002; Young 2009).
Refractive errors, and myopia in particular, are complex genetic traits with a largely unknown etiology. Established environmental factors are education, early reading, and reduced outdoor exposure (Dirani et al. 2009; Ip et al. 2008; McBrien et al. 2008; Morgan and Rose 2005; Rose et al. 2008; Saw et al. 2001; Young 2009). Although heritability estimates are high [50–90 % (Young et al. 2007)], the search for myopia genes is still ongoing. Previous linkage and association studies have led to the identification of at least 18 myopia (MYP) loci, 10 additional chromosomal regions, and several candidate genes (Baird et al. 2010; Young 2009). Replication of these associations has been inconsistent, and their application to the general population is limited (Baird et al. 2010).
Recent genome-wide association studies (GWAS) reported several susceptibility loci for refractive error and myopia (Hysi et al. 2010; Li et al. 2011a, b; Nakanishi et al. 2009; Shi et al. 2011; Solouki et al. 2010). Solouki et al. (2010) and Hysi et al. (2010) were the first to perform a GWAS in a general Caucasian population, and identified susceptibility loci on chromosomes 15q14 and 15q25, respectively. In both studies, carriers of single nucleotide polymorphism (SNP) rs634990 at 15q14 (OR 1.83, 95 % CI 1.42–2.36) and of SNP rs8027411 at 15q25 (OR 1.16, 95 % CI 1.02–1.28) had a higher risk of myopia. Confirmation of these findings was obtained in various replication studies (Hayashi et al. 2011; Hysi et al. 2010; Solouki et al. 2010). However, these replication cohorts were relatively limited in size, increasing the chance of a type 1 error.
To address potential inaccuracies and to investigate generalizability, we investigated the associations between refractive error, and the 15q14 and 15q25 susceptibility loci in a large international replication and meta-analysis study (Consortium of Refractive Error and Myopia, CREAM) including 31 cohorts with various ethnicities from 4 different continents.
Results
Meta-analysis of allelic effects on spherical equivalent (SE)
Complete data on refractive error and genome-wide SNPs were available in all 29 population-based studies comprising 49,364 subjects: 42,224 Caucasians and 7,140 Asians (Table 1; Fig. 1, Supplementary Table 1). This includes the previously reported discovery set consisting of 15,608 (Solouki et al. 2010) and 17,608 subjects (Hysi et al. 2010), respectively.
Table 2 shows the results of the meta-analysis of the 14 SNPs (Hysi et al. 2010; Solouki et al. 2010) at locus 15q14 and 5 SNPs at locus 15q25. The frequency of the effect allele C for top SNP rs634990 at locus 15q14 ranged from 0.38 to 0.64, while frequency of the effect allele A for top SNP rs939661 at 15q25 showed a larger variation, ranging from 0.28 to 0.63 (Supplementary Figure 1). The sample size of each SNP per study is provided in Supplementary Table 1. For locus 15q14, the magnitude and direction of the effects were consistent in all cohorts except Croatia Vis and SIMES. For locus 15q25, there was less consistency; for top SNP rs939661 8 cohorts—both Caucasian and Asian (Australian Twins, Croatia Split, Croatia Vis, EGCUT, FITSA, GHS II, ORCADES, and SIMES)—had a regression beta coefficient in the opposite direction to that of the other studies.
For locus 15q14, the replication set, consisting of all studies except the ones previously used in the discovery analysis, showed a statistically significant association between SE and all SNPs with a best P value 4.53 × 10−14 for top SNP rs634990. Confirmation was achieved in 23 out of 25 Caucasian studies (overall P 3.87 × 10−12 for SNP rs634990), and in 3 out of 4 Asian studies (overall P 2.21 × 10−3 for SNP rs634990). Meta-analysis of the discovery and replication cohorts together provided P value 9.20 × 10−23 for SNP rs634990.
For locus 15q25, neither Caucasian nor Asian validation studies replicated the original association. Meta-analysis of the combined set of the 5 SNPs yielded a lowest P 1.22 × 10−4 for SNP rs939661. As a subsequent analysis, we investigated locus 15q25 in more detail, and tested another 26 SNPs in 26 out of 29 cohorts (no data available in ALSPAC, AREDS 1, and EGCUT). This set of SNPs was not replicated either, however, meta-analysis including the discovery cohort was still significant (best P 2.07 × 10−4 for SNP rs1915726; Supplementary Table 3).
Meta-analysis of risk of myopia for top SNP
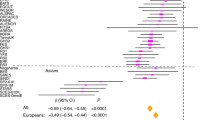
Genotype distributions for rs634990 at locus 15q14 were available for 28 out of 31 studies (all but FITSA, Australian Twins, and SORBS). There was no evidence of heterogeneity in the analyses of homozygote carriers [χ2 21.35 (d.f. 26), P 0.724, I 2 0.0 %] or heterozygote carriers [χ2 24.22 (d.f. 26), P 0.564, I 2 0.0 %]. Therefore, only results from fixed effects meta-analysis were used. Figure 2 shows the forest plots for the risk of myopia for homozygous and heterozygous carriers of the top SNP rs634990. The OR of moderate to high myopia (SE ≤−3 D) versus moderate to high hyperopia (SE ≥+3 D) was 1.88 (95 % CI 1.64, 2.16, P < 0.001) for homozygous carriers of the risk allele at the top SNP rs634990, and 1.33 (95 % CI 1.19, 1.49, P < 0.001) for heterozygous carriers.
Forest plots of odds ratios of myopia (spherical equivalent ≤−3 diopters) versus hyperopia (spherical equivalent ≥+3 diopters) for top SNP rs634990. *For studies without subjects with high or moderate hyperopia, emmetropia was used as a reference group. a Homozygotes carriers of alleles TT versus CC for SNP rs634990. b Heterozygotes carriers of alleles TT versus TC for SNP rs634990
Discussion
Chromosome 15q was first implicated in refractive error and myopia by genome-wide analysis of two large studies located in Northern Europe (Hysi et al. 2010; Solouki et al. 2010). Here, in an international meta-analysis consisting of 31 independent studies from the CREAM consortium, we provide further support that the association with locus 15q14 is robust and present in both Caucasians and Asians. We combined the results with those of the initial study into a powerful meta-analysis of highly associated SNPs with a total study population of 55,177 participants. The combined results showed that all tested SNPs for locus 15q14 were associated with refractive errors, and that homozygous carriers of the top SNP rs634990 had approximately twice the risk of myopia. SNPs at the other locus, 15q25, could not be convincingly replicated.
This study has strengths and limitations. Major strengths of the study include the sample size and the inclusion of different ethnicities. The CREAM consortium represents the largest study on refractive error known to date. Previous replication studies have not been large scaled and focused on populations of the same ancestry (Gao et al. 2012; Lu et al. 2011; Wang et al. 2011). Another advantage of our study is the incorporation of clinical relevant endpoints such as high myopia and high hyperopia. Among the limitations are differences in designs and methods of the studies. (1) Population-based as well as case control studies were incorporated. However, the latter were only two (Kyoto Study and SORBS) and both had results within the same range as the population-based studies. (2) Different types of equipment and measurement methods were used to detect refractive error. These differences are generally subtle, and are not likely to cause false findings. (3) Various methods of genotyping and imputation were used, and genotyping was not complete in all studies. All SNPs at 15q14 had similar effect; thus, we do not think this has influenced these associations. SNPs at 15q25 showed larger variation, and the incomplete genotyping may have underpowered this analysis.
Earlier replication of the 15q14 locus was reported by Hayashi et al. (2011) in a Japanese sample of high myopic probands and controls. In a comparison of 1,125 high myopes (axial length >26.1 mm) versus 1,295 controls, the risk of high myopia was increased for the carriers of the initial top SNP rs634990 [OR 1.84 in homozygotes (95 % CI 1.44–2.36)]. Taken together with the current findings, this suggests that 15q14 plays a role in both common and high myopia.
The 15q14 associated region contains two interesting genes that are both well expressed in the retina, GJD2 and ACTC1. GJD2 encodes the Connexin36 protein, which plays a crucial role in the transmission and processing of visual signals in the retina by enabling intercellular transport of small molecules and ions in photoreceptors, amacrine and bipolar cells (Deans et al. 2002; Guldenagel et al. 2001; Kihara et al. 2009; Striedinger et al. 2005). We speculated that the protein encoded by the other candidate gene, ACTC1, could play a role in scleral remodeling, given the fact that similar actin proteins have been shown to be increased in developing myopic tree shrew eyes (Jobling et al. 2009). Previous GJD2 (Solouki et al. 2010) and ACTC1 (unpublished data) direct sequencing experiments did not reveal a functional variant, but the 15q14 locus appeared to harbor regulatory elements which may influence transcription of these genes (Solouki et al. 2010).
The 15q25 region contains the interesting candidate gene RASGRF1, which is highly expressed in the retina and has previously been implicated in photoreception and visual sensory processes (Fernandez-Medarde et al. 2009; Jones and Moses 2004). The association with this locus and gene is not robust, since none of the initial SNPs replicated significantly, and determination of more SNPs did not increase significance. A type 1 error may explain the initial finding. Another potential cause for the non-replication is a large variation in allele frequencies. The range of allele frequencies at 15q25 (0.28–0.63) was only slightly larger than at 15q14 (0.38–0.64) in our consortium, making this an unlikely explanation (Supplementary Figure 1). Finally, population stratification within cohorts did not appear to play a major role, since only two cohorts had significant principal components, which were addressed in the analyses.
Other GWAS loci were only found for high myopia in Asian case control studies, and they were located on chromosomes 11q24.1 (Nakanishi et al. 2009), 5p15 (Li et al. 2011a), 4q25 (Li et al. 2011b), and 13q12.12 (Shi et al. 2011). The locus on chromosome 5p15 harbors the excellent candidate gene CTNND2 which is involved in retinal morphogenesis, adhesion, retinal cell architecture integrity (Duparc et al. 2006; Paffenholz et al. 1999), and was replicated in subjects of the same ethnicity (Lu et al. 2011). Replication studies for the 4q25 (Gao et al. 2012) and 11q24.1 (Wang et al. 2011) loci were only successful in case of the 4q25 locus; these loci did not have prominent candidate genes.
What should be the next steps? For 15q14, comprehensive resequencing of the entire associated region and the flanking genes can reveal the responsible gene defects which determine the association. Novel techniques such as next-generation sequencing are promising in this regard. Functional studies in knockout animals will shed light on potential protein effects. Finally, evaluation of gene-environment interactions may explain phenotypic variation and help identify high risk groups. For myopia genetics in general, performance of a genome-wide meta-analysis is a logical next step. The current CREAM collaboration is an excellent platform for this project.
In summary, we have convincingly demonstrated that common variants at chromosome 15q14 influence susceptibility for myopia in both Caucasian and Asian populations around the world. Identification of functional variants and responsible genes that explain this association will provide more insight in the complex etiology of myopia.
Materials and methods
Subjects and phenotyping
A total of 31 study cohorts from the Consortium of Refractive Error and Myopia (CREAM) participated in this meta-analysis. 29 population-based as well as 2 case–control studies were included. General methods, descriptives and phenotyping and genotyping methods of the study cohorts can be found in Table 1, the Supplementary Material and Supplementary Table 1, respectively. In short, 22 cohorts consisted of Caucasian, and 5 of Asian study subjects. All studies were performed with the approval of their local Medical Ethics Committee, and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
All studies used a similar protocol for phenotyping. Exclusion criteria were age ≤10 years, and bilateral cataract surgery, laser refractive procedures or other intra-ocular procedures which might alter refraction. Eligible participants underwent a complete ophthalmologic examination including a non-dilated measurement of refractive error (Table 1) of both eyes. Spherical equivalent was calculated according to the standard formula (SE = sphere + ½ cylinder), and the mean of two eyes was used for analysis. When data from only one eye were available, the SE of this eye was used. SE was categorized into low (SE from −1.5 to −3 D), moderate (SE from −3 to −6 D) and high (SE of −6 D or lower) myopia; and also into low (SE from +1.5 to +3 D), moderate (SE from +3 to +6 D) and high (SE of +6 D or higher) hyperopia. Emmetropia was defined as SE equal to or between −1.5 and +1.5 D.
Genotyping and imputation
DNA was extracted according to standard procedures, and genotyping and imputation of SNPs across the entire genome was performed using various methods (Table 1). Samples with a low call rate, with excess autosomal heterozygosity, with sex-mismatch, or outliers identified by the identity-by-state clustering analysis were excluded.
Statistical analysis
Meta-analysis of allelic effects on spherical equivalent
We selected 19 SNPs within loci 15q14 (14 SNPs) and 15q25 (5 SNPs) with a P value of <10−6 from two previous GWAS (Hysi et al. 2010; Solouki et al. 2010). Linear regression models with a 1 degree of freedom trend test were used to examine associations with SE as a quantitative trait outcome, adjusting for age and gender and significant principal components if applicable. From all population-based cohorts, we obtained effect allele, non effect allele, regression coefficient beta, standard error, P value, minor allele and minor allele frequency for each of these SNPs. METAL for Linux was used to perform a meta-analysis on betas and standard errors for all SNPs. First, discovery cohorts (Hysi et al. 2010; Solouki et al. 2010) and replication studies were analyzed separately, followed by a combined meta-analysis. As a second analysis, 26 additional SNPs within the same linkage disequilibrium (LD) block were selected and tested for association using the procedures mentioned above. For these analyses, Bonferroni corrected P values (0.05/number of tested SNPs) of 3.57 × 10−3 for 15q14, and 1.0 × 10−2 (5 SNPs, Table 2) or 1.92 × 10−3 (26 SNPs, Table 3 Supplementary Material) for 15q25 were considered statistically significant.
Meta-analysis of risk of myopia for top SNP
From all population-based and case control studies, we obtained genotype distributions of the replicated top SNPs. We calculated heterogeneity (χ2, I 2 calculated and corresponding P values) between studies, crude OR with corresponding 95 % CI and P value of moderate and high myopia versus moderate and high hyperopia with a random as well as fixed effects meta-analysis using Stata 11. When these analyses provided similar outcomes, data from fixed effect analysis were used. For studies without subjects with high or moderate hyperopia, emmetropia was used as a reference group. A standard P value of <0.05 was considered statistically significant.
References
Baird PN, Schache M, Dirani M (2010) The GEnes in Myopia (GEM) study in understanding the aetiology of refractive errors. Prog Retin Eye Res 29:520–542
Curtin BJ, Karlin DB (1971) Axial length measurements and fundus changes of the myopic eye. Am J Ophthalmol 71:42–53
Deans MR, Volgyi B, Goodenough DA, Bloomfield SA, Paul DL (2002) Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron 36:703–712
Dirani M, Tong L, Gazzard G, Zhang X, Chia A, Young TL, Rose KA, Mitchell P, Saw SM (2009) Outdoor activity and myopia in Singapore teenage children. Br J Ophthalmol 93:997–1000
Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G (2006) Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol 300:647–655
Fernandez-Medarde A, Barhoum R, Riquelme R, Porteros A, Nunez A, de Luis A, de Las Rivas J, de la Villa P, Varela-Nieto I, Santos E (2009) RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J Neurochem 110:641–652
Gao Y, Wang P, Li S, Xiao X, Jia X, Guo X, Zhang Q (2012) Common variants in chromosome 4q25 are associated with myopia in Chinese adults. Ophthalmic Physiol Opt 32:68–73
Guldenagel M, Ammermuller J, Feigenspan A, Teubner B, Degen J, Sohl G, Willecke K, Weiler R (2001) Visual transmission deficits in mice with targeted disruption of the gap junction gene connexin36. J Neurosci 21:6036–6044
Hayashi H, Yamashiro K, Nakanishi H, Nakata I, Kurashige Y, Tsujikawa A, Moriyama M, Ohno-Matsui K, Mochizuki M, Ozaki M, Yamada R, Matsuda F, Yoshimura N (2011) Association of 15q14 and 15q25 with high myopia in Japanese. Invest Ophthalmol Vis Sci 52:4853–4858
He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB (2004) Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci 45:793–799
Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ (2010) A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet 42:902–905
Ip JM, Saw SM, Rose KA, Morgan IG, Kifley A, Wang JJ, Mitchell P (2008) Role of near work in myopia: findings in a sample of Australian school children. Invest Ophthalmol Vis Sci 49:2903–2910
Jobling AI, Gentle A, Metlapally R, McGowan BJ, McBrien NA (2009) Regulation of scleral cell contraction by transforming growth factor-beta and stress: competing roles in myopic eye growth. J Biol Chem 284:2072–2079
Jones C, Moses K (2004) Cell-cycle regulation and cell-type specification in the developing Drosophila compound eye. Semin Cell Dev Biol 15:75–81
Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, Taylor HR, Ikram MK, Congdon NG, O’Colmain BJ, Eye Diseases Prevalence Research G (2004) The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol 122:495–505
Kihara AH, Paschon V, Cardoso CM, Higa GS, Castro LM, Hamassaki DE, Britto LR (2009) Connexin36, an essential element in the rod pathway, is highly expressed in the essentially rodless retina of Gallus gallus. J Comp Neurol 512:651–663
Li YJ, Goh L, Khor CC, Fan Q, Yu M, Han S, Sim X, Ong RT, Wong TY, Vithana EN, Yap E, Nakanishi H, Matsuda F, Ohno-Matsui K, Yoshimura N, Seielstad M, Tai ES, Young TL, Saw SM (2011a) Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology 118:368–375
Li Z, Qu J, Xu X, Zhou X, Zou H, Wang N, Li T, Hu X, Zhao Q, Chen P, Li W, Huang K, Yang J, He Z, Ji J, Wang T, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y (2011b) A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet 20:2861–2868
Lin LL, Shih YF, Hsiao CK, Chen CJ (2004) Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singap 33:27–33
Lu B, Jiang D, Wang P, Gao Y, Sun W, Xiao X, Li S, Jia X, Guo X, Zhang Q (2011) Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci 52:8258–8261
McBrien NA, Gentle A (2003) Role of the sclera in the development and pathological complications of myopia. Prog Retin Eye Res 22:307–338
McBrien NA, Young TL, Pang CP, Hammond C, Baird P, Saw SM, Morgan IG, Mutti DO, Rose KA, Wallman J, Gentle A, Wildsoet CF, Gwiazda J, Schmid KL, Smith E, 3rd, Troilo D, Summers-Rada J, Norton TT, Schaeffel F, Megaw P, Beuerman RW, McFadden SA (2008) Myopia: recent advances in molecular studies; prevalence, progression and risk factors; emmetropization; therapies; optical links; peripheral refraction; sclera and ocular growth; signalling cascades; and animal models. Optom Vis Sci [Epub ahead of print]
Morgan I, Rose K (2005) How genetic is school myopia? Prog Retin Eye Res 24:1–38
Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F (2009) A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet 5:e1000660
Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW (1999) The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res 250:452–464
Rose KA, Morgan IG, Ip J, Kifley A, Huynh S, Smith W, Mitchell P (2008) Outdoor activity reduces the prevalence of myopia in children. Ophthalmology 115:1279–1285
Saw SM (2006) How blinding is pathological myopia? Br J Ophthalmol 90:525–526
Saw SM, Hong CY, Chia KS, Stone RA, Tan D (2001) Nearwork and myopia in young children. Lancet 357:390
Saw SM, Gazzard G, Shih-Yen EC, Chua WH (2005) Myopia and associated pathological complications. Ophthalmic Physiol Opt 25:381–391
Shi Y, Qu J, Zhang D, Zhao P, Zhang Q, Tam PO, Sun L, Zuo X, Zhou X, Xiao X, Hu J, Li Y, Cai L, Liu X, Lu F, Liao S, Chen B, He F, Gong B, Lin H, Ma S, Cheng J, Zhang J, Chen Y, Zhao F, Yang X, Yang C, Lam DS, Li X, Shi F, Wu Z, Lin Y, Yang J, Li S, Ren Y, Xue A, Fan Y, Li D, Pang CP, Zhang X, Yang Z (2011) Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet 88:805–813
Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC (2010) A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet 42:897–901
Striedinger K, Petrasch-Parwez E, Zoidl G, Napirei M, Meier C, Eysel UT, Dermietzel R (2005) Loss of connexin36 increases retinal cell vulnerability to secondary cell loss. Eur J Neurosci 22:605–616
Tano Y (2002) Pathologic myopia: where are we now? Am J Ophthalmol 134:645–660
Vitale S, Ellwein L, Cotch MF, Ferris FL 3rd, Sperduto R (2008) Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol 126:1111–1119
Wang Q, Gao Y, Wang P, Li S, Jia X, Xiao X, Guo X, Zhang Q (2011) Replication study of significant single nucleotide polymorphisms associated with myopia from two genome-wide association studies. Mol Vis 17:3290–3299
Wu HM, Seet B, Yap EP, Saw SM, Lim TH, Chia KS (2001) Does education explain ethnic differences in myopia prevalence? A population-based study of young adult males in Singapore. Optom Vis Sci 78:234–239
Young TL (2009) Molecular genetics of human myopia: an update. Optom Vis Sci 86:E8–E22
Young TL, Metlapally R, Shay AE (2007) Complex trait genetics of refractive error. Arch Ophthalmol 125:38–48
Acknowledgments
We gratefully thank the invaluable contributions of all study participants, their relatives and staff at the recruitment centers. We would like to acknowledge the following agencies and persons:
1958 British Birth Cohort was funded by the Medical Research Council’s Health of the Public grant (PIs Power and Strachan); the Wellcome Trust (083478 to J.S.R.); the National Institute for Health Research as Specialist Biomedical Research Centres partnering respectively with Great Ormond Street and Moorfields Hospitals; and theUlverscroft Vision Research Group.
AGES has been funded by National Institutes of Health (N01AG12100), the National Insitute on Aging and National Eye Institute Intramural Research Programs (ZIAAG007380, ZIAEY000401), Hjartavernd (the Icelandic Heart Association); and the Althingi (the Icelandic Parliament). This study acknowledges the contribution of collaborators on the vision component, Tamara Harris, Lenore Launer, Melissa Garcia, Susan Corwin, Fridbert Jonasson, Johanna Eyrun Sverrisdottir, Sigurdur Sigurdsson, and the staff at Hjartavernd.
Core support for ALSPAC was provided by the UK Medical Research Council (4882); the Wellcome Trust (076467); the University of Bristol; and for this research specifically by the National Eye Research Centre, Bristol (SCIAD053). The study acknowledges Cathy Williams as a guarantor for the contents of this paper.
The AREDS studies were supported by contracts from National Eye Institute/National Institutes of Health, Bethesda, MD, with additional support from Bausch & Lomb Inc, Rochester, NY. The genotyping costs were supported by the National Eye Institute (R01 EY020483 to D.S.) and some of the analyses were supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, USA. AREDS acknowledges Emily Chew and Frederick Ferris, National Eye Institute, National Institutes of Health, Bethesda, MD; and the Center for Inherited Disease Research, Baltimore, MD where SNP genotyping was carried out.
KORA would like to acknowledge Christian Geiger and the Center for Inherited Disease Research, Baltimore, MD where SNP genotyping was carried out.
Australian Twins was supported by an Australian National Health and Medical Research Council (NHMRC) Enabling Grant (2004-2009, 350415, 2005-2007); Clifford Craig Medical Research Trust; Ophthalmic Research Institute of Australia; American Health Assistance Foundation; Peggy and Leslie Cranbourne Foundation; Foundation for Children; Jack Brockhoff Foundation; National Institutes of Health/National Eye Institute (RO1EY01824601 (2007-2010)); Pfizer Australia Senior Research Fellowship (to D.A.M.); and Australian NHMRC Career Development Award (to S.M.). Genotyping was funded by an NHMRC Medical Genomics Grant and NIH Center for Inherited Disease Research as part of an National Eye Institute National Institutes of Health project grant, Australian sample imputation analyses were carried out on the Genetic Cluster Computer which is financially supported by the Netherlands Scientific Organization (NWO48005003). Australian Twins thanks Stuart Macgregor, Grant W. Montgomery, Nicholas G. Martin, Scott D. Gordon, Dale R. Nyholt, Sarah E. Medland, Brian P. McEvoy, Margaret J. Wright, Anjali K. Henders, Megan J. Campbell for ascertaining and processing genotyping data; Jane MacKinnon, Shayne Brown, Lisa Kearns, Jonathan Ruddle, Paul Sanfilippo, Sandra Staffieri, Olivia Bigault, Colleen Wilkinson, Jamie Craig, Yaling Ma, Julie Barbour for assisting with clinical examinations; and Dr Camilla Day and staff.
The Blue Mountains Eye Study was supported by the Australian National Health & Medical Research Council (NH&MRC), Canberra Australia (974159, 211069, 457349, 512423, 475604, 529912); the Centre for Clinical Research Excellence in Translational Clinical Research in Eye Diseases; NH&MRC research fellowships (358702, 632909 to J.J.W, 1028444 to P.N.B.); and the Wellcome Trust, UK as part of Wellcome Trust Case Control Consortium 2 (A Viswanathan, P McGuffin, P Mitchell, F Topouzis, P Foster) for genotyping costs of the entire BMES population (085475B08Z, 08547508Z, 076113). The Centre for Eye Research Australia receives Operational Infrastructure Support from the Victorian government. BMES acknowledges Elena Rochtchina from the Centre for Vision Research, Department of Ophthalmology and Westmead Millennium Institute University of Sydney (NSW Australia); John Attia, Rodney Scott, Elizabeth G. Holliday from the University of Newcastle (Newcastle, NSW Australia); Jing Xie, Maria Schache and Andrea J. Richardson from the Centre for Eye Research Australia, Department of Ophthalmology, University of Melbourne; Michael Inouye, The Walter and Elisa Hall Institute of Medical Research (Victoria, Australia); Ananth Viswanathan, Moorfields Eye Hospital (London, UK); Paul J. Foster, NIHR Biomedical Research Centre for Ophthalmology, UCL Institute of Ophthalmology & Moorfields Eye Hospital (London); Peter McGuffin, MRC Social Genetic and Developmental Psychiatry Research Centre, Institute of Psychiatry, King’s College (London, United Kingdom); Fotis Topouzis, Department of Ophthalmology, School of Medicine, Aristotle University of Thessaloniki, AHEPA Hospital (Thessaloniki, Greece); Xueling Sim, National University of Singapore; members of the Wellcome Trust Case Control Consortium 2. (Membership of Wellcome Trust Case Control Consortium 2 Peter Donnelly1,2, Ines Barroso3, Jenefer M Blackwell4, 5, Elvira Bramon6, Matthew A Brown7, Juan P Casas8, Aiden Corvin9, Panos Deloukas3, Audrey Duncanson10, Janusz Jankowski11, Hugh S Markus12, Christopher G Mathew13, Colin NA Palmer14, Robert Plomin15, Anna Rautanen1, Stephen J Sawcer16, Richard C Trembath13, Ananth C Viswanathan17, Nicholas W Wood18, Chris C A Spencer1, Gavin Band1, Céline Bellenguez1, Colin Freeman1, Garrett Hellenthal1, Eleni Giannoulatou1, Matti Pirinen1, Richard Pearson1, Amy Strange1, Zhan Su1, Damjan Vukcevic1, Cordelia Langford3, Sarah E Hunt3, Sarah Edkins3, Rhian Gwilliam3, Hannah Blackburn3, Suzannah J Bumpstead3, Serge Dronov3, Matthew Gillman3, Emma Gray3, Naomi Hammond3, Alagurevathi Jayakumar3, Owen T McCann3, Jennifer Liddle3, Simon C Potter3, Radhi Ravindrarajah3, Michelle Ricketts3, Matthew Waller3, Paul Weston3, Sara Widaa3, Pamela Whittaker3 1 Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7LJ, UK; 2 Dept Statistics, University of Oxford, Oxford OX1 3TG, UK; 3 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK; 4 Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, 100 Roberts Road, Subiaco, Western Australia 6008; 5 Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge CB2 0XY, UK; 6 Department of Psychosis Studies, NIHR Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King’s College London and The South London and Maudsley NHS Foundation Trust, Denmark Hill, London SE5 8AF, UK; 7 University of Queensland Diamantina Institute, Brisbane, Queensland, Australia; 8 Dept Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London WC1E 7HT and Dept Epidemiology and Public Health, University College London WC1E 6BT, UK; 9 Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin 2, Eire; 10 Molecular and Physiological Sciences, The Wellcome Trust, London NW1 2BE; 11 Centre for Digestive Diseases, Queen Mary University of London, London E1 2AD, UK and Digestive Diseases Centre, Leicester Royal Infirmary, Leicester LE7 7HH, UK and Department of Clinical Pharmacology, Old Road Campus, University of Oxford, Oxford OX3 7DQ, UK; 12 Clinical Neurosciences, St George’s University of London, London SW17 0RE; 13 King’s College London Dept Medical and Molecular Genetics, School of Medicine, Guy’s Hospital, London SE1 9RT, UK; 14 Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee DD1 9SY, UK; 15 King’s College London Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Denmark Hill, London SE5 8AF, UK; 16 University of Cambridge Dept Clinical Neurosciences, Addenbrooke’s Hospital, Cambridge CB2 0QQ, UK; 17 NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1 V 2PD, UK; 18 Dept Molecular Neuroscience, Institute of Neurology, Queen Square, London WC1 N 3BG, UK.).
The CROATIA studies were funded by grants from the Medical Research Council (UK) and from the Republic of Croatia Ministry of Science, Education and Sports (10810803150302); and the CROATIA-Korcula genotyping was funded by the European Union framework program 6 project EUROSPAN (LSHGCT2006018947). The CROATIA studies acknowledges Dr. Goran Bencic, Prof. Zoran Vatavuk, Biljana Andrijević Derk, Valentina Lacmanović Lončar, Krešimir Mandić, Antonija Mandić, Ivan Škegro, Jasna Pavičić Astaloš, Ivana Merc, Miljenka Martinović, Petra Kralj, Tamara Knežević and Katja Barać-Juretić as well as the recruitment team from the Croatian Centre for Global Health, University of Split and the Institute of Anthropological Research in Zagreb for the ophthalmological data collection; Peter Lichner and the Helmholtz Zentrum Munchen (Munich, Germany), AROS Applied Biotechnology, Aarhus, Denmark and the Wellcome Trust Clinical facility (Edinburgh, United Kingdom) for the SNP genotyping all studies.
ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the Medical Research Council Human Genetics Unit and the European Union framework program 6 EUROSPAN project (LSHGCT2006018947). ORCADES acknowledges the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, in particular Margaret Pratt who performed the eye measurements,as well as the administrative team in Edinburgh University; and the Wellcome Trust Clinical facility (Edinburgh, United Kingdom) for DNA extraction; and Peter Lichner and the Helmholtz Zentrum Munchen (Munich, Germany) for genotyping.
EGCUT received financing by FP7 grants (201413, 245536); Estonian Government (SF0180142s08); and the European Union through the European Regional Development Fund, in the frame of Centre of Excellence in Genomics and Estonian Research Infrastructure’s Roadmap. EGCUT acknowledges Ms. M. Hass and Mr. V. Soo.
FITSA was supported by ENGAGE (FP7-HEALTH-F4-2007, 201413); the Academy of Finland Center of Excellence in Complex Disease Genetics (213506, 129680); the Academy of Finland Ageing Programme; and the Finnish Ministry of Culture and Education and University of Jyväskylä. For FITSA the contributions of Emmi Tikkanen, Samuli Ripatti and Jaakko Kaprio are acknowledged.
Framingham Eye Study was supported by NEI (N01EY22112, N01EY92109); the National Heart, Lung, and Blood Institute (N02HL64278) for SHARe genotyping; Boston University (N01HC25195); and by intramural funds of the National Human Genome Research Institute, NIH, USA (to R.W. and J.E.B.W.) GHS was funded through the government of Rheinland-Pfalz (“Stiftung Rheinland Pfalz für Innovation” (AZ961386261733); the research programs “Wissen schafft Zukunft” and “Schwerpunkt Vaskuläre Prävention” of the Johannes Gutenberg-University of Mainz; Boehringer Ingelheim; PHILIPS Medical Systems; National Genome Network “NGFNplus” by the Federal Ministry of Education and Research, Germany (A301GS0833).
KORA was financed by the Helmholtz Center Munich, German Research Center for Environmental Health; the German Federal Ministry of Education and Research; the State of Bavaria; the German National Genome Research Network (NGFN-2 and NGFNPlus) (01GS0823); Munich Center of Health Sciences as part of LMUinnovativ; the German Research Counsil (DFG) (WI182041 to K.O.); the genotyping costs were supported by the National Eye Institute (R01 EY020483 to D.S.) and some of the analyses were supported by the Intramural Research Program of the National Human Genome.
The Kyoto Study was supported by the Japan Society for the Promotion of Science, Tokyo (21249084, 22791653).
MESA and MESA SNP Health Association Resource (SHARe) are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) (N01HC95159, N01HC95169, RR-024156, N02HL64278 (SHARe genotyping)); the National Institutes of Health (Intramural Research Program of the National Eye Institute, (ZIAEY000403); (R01HL071205 to MESA Family); the Clinical Translational Science Institute (UL1RR033176); and the Cedars-Sinai General Clinical Research Center (RR00425). MESA thanks all investigators, especially Drs. Mary Frances Cotch, Jerome I. Rotter, Ronald Klein, and Tien Y. Wong in the Eye Working Group, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
OGP Talana was supported by grants from the Italian Ministry of Education, University and Research (5571DSPAR2002, 718Ric2005). OGP Talana thanks the Ogliastra population and the municipal administrators for their collaboration to the project and for economic and logistic support.
The Rotterdam Study and ERF were supported by the Netherlands Organisation of Scientific Research (NWO) (Vidi 91796357); Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands; Netherlands Organization for Health Research and Development (ZonMw); UitZicht; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); the Municipality of Rotterdam; the Netherlands Genomics Initiative/NWO; Center for Medical Systems Biology of NGI; Lijf en Leven; M.D. Fonds; Henkes Stichting; Stichting Nederlands Oogheelkundig Onderzoek; Swart van Essen; Bevordering van Volkskracht; Blindenhulp; Landelijke Stichting voor Blinden en Slechtzienden; Rotterdamse Vereniging voor Blindenbelangen; OOG; Algemene Nederlandse Vereniging ter Voorkoming van Blindheid; the Rotterdam Eye Hospital Research Foundation; and Topcon Europe. Rotterdam Study and ERF thank Ada Hooghart, Corina Brussee, Riet Bernaerts-Biskop, Patricia van Hilten, Pascal Arp, Jeanette Vergeer, Marijn Verkerk and Sander Bervoets.
The Singapore studies (SCORM, SP2, SiMES, SINDI) were supported by the National Medical Research Council, Singapore (NMRC 07962003, NMRC 11762008), Singapore Bio-Medical Research Council (0612119466, 0913519616).
The Sorbs study was supported by the Interdisciplinary Centre for Clinical Research at the University of Leipzig (B27 to A.T.) from the German Diabetes Association (to A.T.); the DHFD, Diabetes Hilfs- und Forschungsfonds Deutschland (to A.T.); the European Commission under a Marie Curie Intra-European Fellowship (to R.M.); the European Community's Seventh Framework Programme (FP720072013); and ENGAGE project (HEALTHF42007201413). We thank Michael Stumvoll and Peter Kovacs for the excellent project coordination and fruitful discussion, furthermore Knut Krohn (Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig) for the genotyping support.
TwinsUK received funding from the Wellcome Trust; the European Union MyEuropia Marie Curie Research Training Network; Guide Dogs for the Blind Association; the European Community’s FP7 (HEALTHF22008201865GEFOS); ENGAGE (HEALTHF42007201413); the FP-5 GenomEUtwin Project (QLG2CT200201254); US National Institutes of Health/National Eye Institute (1RO1EY018246); NIH Center for Inherited Disease Research; the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ National Health Service Foundation Trust partnering with King’s College London.
The Young Finns Study was financially supported by the Academy of Finland (134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi)); the Social Insurance Institution of Finland, Kuopio, Tampere; Turku University Hospital Medical Funds (grant 9M048 to T.L.); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation; Tampere Tuberculosis Foundation; and Emil Aaltonen Foundation (to T.L.).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments comply with the current laws in which they were performed.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Verhoeven, V.J.M., Hysi, P.G., Saw, SM. et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet 131, 1467–1480 (2012). https://doi.org/10.1007/s00439-012-1176-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-012-1176-0