Abstract
To confirm and refine associations of human leukocyte antigen (HLA) genotypes with variable antibody (Ab) responses to hepatitis B vaccination, we have analyzed 255 HIV-1 seropositive (HIV+) youth and 80 HIV-1 seronegatives (HIV−) enrolled into prospective studies. In univariate analyses that focused on HLA-DRB1, -DQA1, and -DQB1 alleles and haplotypes, the DRB1*03 allele group and DRB1*0701 were negatively associated with the responder phenotype (serum Ab concentration ≥ 10 mIU/mL) (P = 0.026 and 0.043, respectively). Collectively, DRB1*03 and DRB1*0701 were found in 42 (53.8%) out of 78 non-responders (serum Ab <10 mIU/mL), 65 (40.6%) out of 160 medium responders (serum Ab 10–1,000 mIU/mL), and 27 (27.8%) out of 97 high responders (serum Ab >1,000 mIU/mL) (P < 0.001 for trend). Meanwhile, DRB1*08 was positively associated with the responder phenotype (P = 0.010), mostly due to DRB1*0804 (P = 0.008). These immunogenetic relationships were all independent of non-genetic factors, including HIV-1 infection status and immunodeficiency. Alternative analyses confined to HIV+ youth or Hispanic youth led to similar findings. In contrast, analyses of more than 80 non-coding, single nucleotide polymorphisms within and beyond the three HLA class II genes revealed no clear associations. Overall, several HLA-DRB1 alleles were major predictors of differential Ab responses to hepatitis B vaccination in youth, suggesting that T-helper cell-dependent pathways mediated through HLA class II antigen presentation are critical to effective immune response to recombinant vaccines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) infection affects 350–400 million people globally (Custer et al. 2004; Dienstag 2008; Fattovich et al. 2008; Lai and Yuen 2008; McGovern 2007). Vaccines based on recombinant HBV surface antigen (HBsAg) offer a safe and effective strategy for preventing HBV infection, as anti-HBsAg antibody (Ab) concentrations can exceed the minimum protective level of 10 mIU/mL for up to 95% of healthy recipients. Nonetheless, lack of efficacy can be a problem to vaccine recipients with compromised or impaired immune system (Collier et al. 1988; Zuin et al. 1992), which has prompted the search for further improvements through (a) increasing the amount of antigen given in each injection (Cornejo-Juarez et al. 2006; Cruciani et al. 2009; de Vries-Sluijs et al. 2008; Fonseca et al. 2005; Pasricha et al. 2006), (b) adding an extra (boosting) dose when the standard three-dose regimen fails to induce protective Ab response (Cruciani et al. 2009; Rey et al. 2000), and (c) using immunoregulatory cytokine or unmethylated CpG oligonucleotide as an adjuvant in one of the priming doses (Cooper et al. 2008; Sasaki et al. 2003).
Suboptimal Ab response to full-dose HBV vaccination was common in adolescents and youth enrolled by the Reaching for Excellence in Adolescent Care and Health (REACH) study supported by the Adolescent Medicine Trials Network for HIV/AIDS Interventions (ATN) (Wang et al. 2004; Wilson et al. 2001). Key factors associated with this unfavorable outcome included: (1) immunodeficiency as a result of HIV-1 infection, (2) human leukocyte antigen (HLA) class II genotype, especially DRB1*07, and (3) promoter sequence variations in several cytokine genes (Wang et al. 2004). To confirm and refine some of these findings, new interventional studies nested within ATN have enrolled youth for prospective evaluation of HBV vaccination. These studies have facilitated the systematic analyses of HLA class II alleles and haplotypes in relation to differential Ab responses to HBV vaccination. Genotypes defined by non-coding, single nucleotide polymorphisms (SNPs) within and beyond the HLA class II region can also be tested for their potential associations.
Methods
Study populations
Between February 2006 and June 2008, 255 HIV-1 seropositive (HIV+) youth (12–24 years of age) and 80 HIV-1 seronegatives (HIV−) (12–17 years of age) were enrolled at 22 clinical sites (listed in Appendix) into prospective studies (http://www.clinicaltrials.gov/ct/show/NCT00142740) (Table 1). The HIV− subjects were recruited from the US and Puerto Rico, while the HIV+ youth came from US, Puerto Rico, Brazil, and South Africa. The primary objective was to evaluate genetic influences on peak Ab responses in patients with three full doses of hepatitis B vaccination, based on analyses of eligible patients who were free of hepatitis before completing full vaccination regimens within the first 24 weeks (6 months) after enrollment. For HIV+ youth, their baseline CD4+ T cell counts were also measured as potential covariates for immune responses. This study was first approved by the Institutional Review Board (IRB) at University of Alabama at Birmingham (UAB), as protocol F051212004. Additional IRB approval was obtained from each participating clinical site before patients were asked to provide written informed consent for participation. Import of patient specimens from international sites in Brazil and South Africa to US was approved by the Centers for Disease Control and Prevention (permit no. 2006-12-053 and 2007-12-015).
Quantification and categorization of serum Ab concentration
Using serum sample drawn at week 28, i.e., 4 weeks after the final recommended dose of vaccine, Ab specific for HBsAg was measured in a central laboratory (Quest Diagnostics Inc., Baltimore, MD), using two commercial assays that differ in their lower limits of detection (LLD) (3 and 5 mIU/mL). The vast majority (98.5%) of samples were tested using the second assay, which provided an upper limit of detection (ULD) at 1,000 mIU/mL. A small number of subjects had no remaining serum to allow re-measure and so these subjects only had Ab results reported using the first test (ULD at 150 mIU/mL). Patients with peak serum Ab concentration less than 10 mIU/mL (the minimum protection level) were classified as non-responders (NR). Those with Ab concentrations ranging from 10 to 1,000 mIU/mL were designated as medium responders (MR), while high responders (HR) were the remainder of patients with Ab concentrations over 1,000 mIU/mL.
PCR-based genotyping of three HLA class II genes (HLA-DRB1, -DQA1, and -DQB1) in the human major histocompatibility complex (MHC)
Using DNA extracted from whole blood or buffy coat (PureGen kits from Gentra/Qiagen, Valencia, CA, USA), HLA-DQA1 and -DQB1 alleles were resolved with automated hybridization of sequence-specific oligonucleotide (SSO) probes to gene-specific PCR amplicons (Innogenetics Inc., Alphartetta, GA). Tests were facilitated by the Auto LiPA 30 system, under conditions recommended by the manufacturer (Innogenetics). A total of 35 and 37 SSO probes were used for HLA-DQA1 and -DQB1 typing, respectively. Homozygosity was confirmed and ambiguities with SSO results were resolved by sequencing-based typing (SBT), using commercial kits from Abbott Molecular Inc. (Des Plaines, IL). HLA-DRB1 genotyping began with SBT (Abbott Molecular Inc.), with ambiguities resolved by SSO assays (57 probes) (Innogenetics). The same or similar genotyping techniques had been applied extensively in earlier studies involving samples from African, European, and North American populations (Shao et al. 2004; Tang et al. 2004, 2008). The final allele assignments for the three HLA class II genes considered two major functional properties. First, allele groups defined by the first two-digit designations (e.g., DRB1*01) often correspond to serological specificities (as alloantigens) (Schreuder et al. 2005) or shared motifs and lineages (Agrawal et al. 2007; Bergstrom et al. 1999; Dorak et al. 2006; Hohjoh et al. 2003). Second, alleles with four-digit designations (e.g., DRB1*0101, *0102, and *0103) define functional units that differ in amino acid sequences and the protein structure. Higher (six-digit) resolution like DRB1*010201 was also achievable for various alleles in the study population, with the 5th and 6th digits capturing additional, synonymous DNA polymorphisms. However, only DRB1*1101 had multiple common alleles with different six-digit designations (i.e., *110101 and *110102) in the study population. Further intronic sequence polymorphisms reflected by the 7th and 8th digits were not considered because intronic sequences were not part of our HLA genotyping strategies and because only a small number of alleles have such known designations (Robinson et al. 2006).
Computational assignment of HLA class II haplotypes
Following resolution of individual HLA alleles at each locus, linkage disequilibrium (LD) between specific HLA allele groups (two-digit designations) and alleles (four-digit designations) at neighboring loci were determined by relative difference (D′) between the observed and expected frequencies of co-existence (presence) on the same chromosome or by correlation coefficients (r). Two-locus haplotype assignments were based on the expectation-maximization (EM) algorithm in SAS Genetics (SAS Institute, Cary, NC), which was further applied to the evaluation of extended, three-locus haplotypes. All haplotypes assigned computationally were unambiguous, with statistical probability exceeding 99%.
Primary association analyses of HLA class II variants
Using SAS program package, version 9.2 (including SAS Genetics) (SAS Institute), the distribution of HLA class II alleles and haplotypes was first assessed for Hardy–Weinberg equilibrium (HWE) among patient groups defined by race, ethnicity, HIV-1 infection status, and Ab responses (NR, MR, and HR). Population (marker) frequencies of HLA allele groups, alleles, and their haplotypes were then compared across the three groups of patients defined by Ab responses. Individual variants with nominal P < 0.050 in univariate tests for trend (logit procedure) were further evaluated in multivariable logistic regression models. Different model selection procedures (backward, forward, stepwise) were applied until a reduced model could sufficiently capture all independent predictors of the responder phenotype. The summary statistical measures included effective sample sizes as well as strengths of associations, as indicated by beta estimates (mean ± standard error), proportional odds ratio (pOR) across three Ab response groups (HR, MR, and NR), OR for being responders (HR + MR vs. NR), 95% confidence interval (CI) for all pOR and OR estimates, along with P values (either nominal or adjusted).
Genotyping of SNPs within and beyond the HLA class II region
Single nucleotide polymorphism selection focused on three major genomic blocks within and between HLA class I and class II genes (Table S1). The first (HLA class I region) has three genes (ZNRD1, HLA-C, and HCP5) recently implicated as important to HIV-1 pathogenesis in several genome-wide association studies (Dalmasso et al. 2008; Fellay et al. 2007; Limou et al. 2009) and additional follow-up studies (Catano et al. 2008; Shrestha et al. 2009; van Manen et al. 2009). The second (central HLA region) has a cluster of genes (TNF, LTA, and LTB) encoding tumor necrosis factor alpha (TNF-α), lymphotoxin alpha (LT-α or TNF-β), and LT-β that regulate inflammatory responses. The third (HLA class II region) has multiple non-coding SNPs that can effectively tag HLA class II alleles associated with autoimmune and related diseases (de Bakker et al. 2006). Gene-specific SNPs in HLA class I and central regions were selected in five steps. First, SNPs assembled through SNPper (http://snpper.chip.org/bio/snpper-explain) were filtered to allow an initial focus on “functional SNPs” with a potential phenotypic effect, including those located in exons, transcription factor binding sites, and intron/exon boundaries. Second, SNPs with minor allele frequencies (MAF) <0.02 in any of the three major populations (Africans, Asians, and Europeans) targeted by the International HapMap Project (http://www.hapmap.org/cgi-perl/gbrowse/hapmap26_B36/) were excluded for lack of statistical power. Third, the Tagger program (http://www.broad.mit.edu/mpg/tagger/) was applied to select additional tagging SNPs (tagSNPs) that would capture multiple SNPs using a stringent tagging algorithm (r 2 ≥ 0.8), with the r 2 values derived from the HapMap project or the Perlegen Genotype Browser (http://genome.perlegen.com/browser/index.html). Fourth, SNPs commonly analyzed in earlier population studies (de Bakker et al. 2006; Fellay et al. 2007) were added to serve as benchmarks that could facilitate inter-cohort comparisons. Fifth, SNPs chosen after the first four steps were evaluated for their respective suitability for bead array-based genotyping (Goldengate SNP typing assay) at Illumina (San Diego, CA). SNPs predicted to have low (<50%) success rates were replaced by new ones of equal tagging capabilities. The last (fifth) step was repeated twice to yield a total of 88 (3 coding and 85 non-coding) SNPs (Table S1) for high-throughput genotyping, with quality control facilitated by previously genotyped reference DNA and by random duplicates of DNA samples derived from the study population.
Statistical approach to analyzing SNP genotypes
For each of the 88 MHC SNPs analyzed here, homozygous and heterozygous genotypes were tested in 3 × 3 and 3 × 2 contingency tables, i.e., patients with (2 or 1 copy) and without (0 copy) the minor allele at each locus were stratified by the three levels of Ab response (NR, MR, and HR). A nominal P value < 0.050 in Chi-square test was considered indicative of possible associations and all neighboring SNPs were evaluated jointly for potential involvement of local haplotypes. Evaluation of the relative and independent contribution of SNP alleles and haplotypes required statistical adjustments for HLA and non-genetic factors revealed by primary association analyses. Given the multiple testing of 88 target SNPs, about four random (false positive) discoveries were anticipated when type I statistical error was accepted at the 0.050 level, while a truly probable association would require a P value ≤5.7 × 10−4 to withstand correction for multiple comparisons. As a compromise between least and most stringent evaluations, formal multivariable statistical models were considered useful only for SNP variants with P < 0.001 in the initial univariate tests.
Results
Patient characteristics and assessment of non-genetic factors in Ab responses
Among the 255 HIV-1 seropositive (HIV+) and 80 seronegative (HIV−) youth enrolled into this study, HIV− individuals had a mean age of 15.8 years and they were mostly male (63.8%) and Hispanic (75.0%). HIV+ individuals were slightly older (average age = 20.9 years, P < 0.0001 when compared with HIV− subjects) and they were mostly female (61.6%) and Hispanic (66.3%).
As seen in analyses of another youth population (Wang et al. 2004; Wilson et al. 2001), Ab response clearly differed between HIV+ and HIV− recipients (Table 1). More specifically, 5.0% of HIV− subjects were classified as NR (Ab concentration <10 mIU/mL), 48.8% as MR (10–1,000 mIU/mL), and 46.3% as HR (>1,000 mIU/mL), while HIV+ subjects had 29.0% NR, 47.5% MR, and 23.5% HR (overall P < 0.0001 by logistic regression). In univariate analysis, the negative impact of HIV-1 infection was clearly reflected by the mean parameter (β) estimate and standard error (SE) (−0.61 ± 0.13, P < 0.0001), along with the estimates of proportional odds ratio (pOR = 0.29) and 95% confidence interval (CI) (0.18–0.48) in test for trend across the three patient groups defined by high to low Ab responses (i.e., ordered as HR, MR, and NR) (Table 2). To minimize potential confounding by non-genetic factors, all subsequent association analyses considered age, sex, ethnicity, and HIV-1 infection status as cofactors (whenever possible).
Distribution and LD of HLA class II variants
Before any stratification, common allele groups found in at least ten individuals ranged from five at HLA-DQA1 and -DQB1 loci to 12 at the HLA-DRB1 locus. These allele groups contributed to 18 common DRB1–DQB1 haplotypes. Distribution of homozygous and heterozygous genotypes at each locus did not deviate from HWE, even for subsets of HLA typing results restricted to either racial groups or HIV-1 infection status (data not shown). Similarly, distribution of four-digit alleles also conformed to HWE, and the common alleles detected at each locus ranged from 12 at the HLA-DQA1 locus to 25 at the HLA-DRB1 locus. Twenty DRB1–DQB1 haplotypes were also common in tabulations based on the four-digit allelic designations.
LD was quite strong for many four-digit alleles resolved at the three HLA class II loci, leading to the unambiguous (probability >99%) assignment of three-locus haplotypes in all 335 individuals. However, within the three main racial groups (Table 1), pairs of common alleles with strong enough LD (r 2 ≥ 0.80) to be mutually tagging were limited to (a) DRB1*0301 versus DQB1*0201 (r = 0.82–0.95), (b) DRB1*0301 versus DQA1*0501 (r = 0.92–1.00), (c) DRB1*0701 versus DQA1*0201 (r = 0.86–0.98), (d) DRB1*1101 versus DQA1*0105 (r = 0.72–0.94), (e) DRB1*1301 versus DQB1*0603 (r = 0.81–0.92), (f) DRB1*1401 versus DQA1*0104 (r = 0.92–1.00), (g) DRB1*1401 versus DQB1*0503 (r = 0.92–1.00), (h) DRB1*1402 versus DQB1*0503 (r = 0.89–0.92), (i) DRB1*1502 versus DQB1*0601 (r = 0.71–1.00), (j) DQA1*0501 versus DQB1*0201 (r = 0.83–0.96), and (k) DRB1*0301–DQB1*0201 haplotype versus DQA1*0501 (r = 0.92–1.00). These allele combinations and their two-digit allele groups were treated as joint entities in order to minimize the number of tests for genetic associations with variable Ab responses.
As HIV-1 infection status alone was a strong predictor of suboptimal Ab response, HLA class II alleles and haplotypes that were differentially (P < 0.050) distributed among HIV+ and HIV− subjects were also sorted (Table S2). Alleles enriched in HIV− subjects were DRB1*0407, DRB1*0802, and DQA1*0301. Alleles enriched in HIV+ subjects were DRB1*0804, DQA1*0103, DQB1*0503, and DQB1*0603 (Table S2).
Individual HLA class II variants in relation to differential Ab responses in the entire study population
Primary association analyses tested the key hypothesis that HLA class II alleles and/or haplotypes, especially those observed in another youth population (Wang et al. 2004), correlate with variable Ab response to hepatitis B vaccination, irrespective of other characteristics of the study population. After accounting for sample size (statistical power) and patterns of LD (genetic confounding), four allele groups stood out to be the most likely predictors of differential Ab responses (Table 2). First, DRB1*03 (mostly *0301 and *0302) and DRB1*07 (exclusively *0701) were negatively associated with the responder phenotype, with similar effective sample size (n = 71 for both), β estimates (−0.56 for both) and P values (≤0.043), as well as similar pOR (≤ 0.60). Second, DRB1*08 seen in 51 patients had a positive association (β = 0.75, P = 0.010; pOR = 2.11, 95% CI = 1.20–3.72), mostly due to DRB1*0804 (n = 21, β = 1.17 ± 0.44, P = 0.008; pOR = 3.22, 95% CI = 1.35–7.63). In contrast, DRB1*0802 (Table S2) as another common allele in the DRB1*08 group did not show a clear trend for positive or negative association (n = 16, β = 0.18 ± 0.48, P > 0.70), neither did the less frequent allele DRB1*0801 (n = 9, β = 0.74 ± 0.64, P = 0.249). Third, DQB1*02, which was found in 116 patients and in LD with DRB1*03 (r = 0.32–0.43, P ≤ 0.0001) and DRB1*07 (r = 0.55–0.72, P ≤ 8.0 × 10−10), paralleled the associations of the two DRB1 allele, with similar parameter estimates (β = −0.50 ± 0.22, P = 0.020; pOR = 0.60, 95% CI = 0.40–0.93).
In multivariable models, the impact of three HLA-DRB1 variants on Ab responses was clearly independent of one another and these associations were insensitive to statistical adjustments for the effects of HIV-1 infection and other potential confounders (adjusted P = 0.005–0.040) (Table 2). Alternating DRB1*03 and DRB1*0701 with DQB1*02 did not change the parameter estimates for non-genetic factors and DRB1*0804, but it was evident that DQB1*02 itself was not a statistically significant factor (adjusted P = 0.100) (Table 2). Collectively, DRB1*03 and DRB1*0701 were found in 42 (53.8%) out of 78 NR, 65 (40.6%) out of 160 MR, and 27 (27.8%) out of 97 HR (P < 0.001 for trend).
Evaluation of HLA haplotypes in the entire study population
Associations attributed to individual DRB1 alleles or allele groups were partially reflected by several DRB1-DQB1 haplotypes (Table S2). For example, the negative impact of DRB1*03 and DRB1*07 on Ab response was mostly due to the DRB1*03–DQB1*04 (DRB1*0302–DQB1*0402) haplotype (β = −0.75 ± 0.41, P = 0.067) and the DRB1*07–DQB1*02 (DRB1*0701–DQB1*0202) haplotype (β = −0.48 ± 0.26, P = 0.066), respectively. The positive association of DRB1*08 was mainly because of DRB1*08–DQB1*03 (n = 22, β = 1.17 ± 0.43, P = 0.007) or DRB1*0804–DQB1*0301 (n = 18, β = 1.26 ± 0.48, P = 0.009), but not DQB1*0301 itself (n = 108, β = 0.23 ± 0.22, P = 0.290). The parameter estimates for the major haplotypes involving DRB1*08 were highly comparable with those for DRB1*0804 alone (Table 2). Further analyses of extended (three-locus) haplotypes did not yield any definitive relationships.
Two alternative models to allow inter-study comparisons
To facilitate direct comparison with earlier findings based on vaccine recipients without apparent immunodeficiency, 27 HIV+ subjects with severe immunodeficiency (<200 CD4+ T cells/μL) were excluded from the testing of an alternative model (Table 3). DRB1*03, DRB1*0701, and DRB1*0804 remained as cofactors associated with differential Ab responses (P = 0.002–0.006), even after statistical adjustment for residual effects of HIV-1 infection and other non-genetic factors. A second alternative model used the strategy tested in another cohort of youth (Wang et al. 2004), when responders were not further differentiated into HR and MR groups. Again, DRB1*03 and *0701 were independently associated with differential Ab response status (Table 3), with similar parameter estimates (β = −0.88 ± 0.35 and −1.03 ± 0.36, P = 0.013 and 0.005, respectively). The effect of DRB1*0804 could not be reliably established, as the allele was only present in the aggregated responder group (HR + MR).
Analyses restricted to 255 HIV+ subjects
Stepwise analyses were further applied to the 255 HIV+ individuals alone (Table 4). By univariate analyses, severe immunodeficiency (<200 CD4+ T cells/μL) and moderate immunodeficiency (200–450 CD4+ T cells/μL) had a strong negative impact on Ab responses (P < 0.001). DRB1*0701 became a minor factor (n = 56, P = 0.159) when compared with DRB1*03 (n = 58, P = 0.039) and DRB1*0804 (n = 18, P = 0.003). Two additional DRB1 variants, DRB1*04 (n = 44, P = 0.054) and DRB1*15 (n = 55, P = 0.092), were recognized as marginally unfavorable and favorable, respectively, with the latter found exclusively on the DRB1*15–DQB1*06 haplotype. Only DRB1*03, DRB1*0701, and DRB1*0804 could be retained as independent factors (adjusted P = 0.005–0.029) in the reduced multivariable model, after statistical adjustments for the clear, negative impact of severe and moderate immunodeficiency (P < 0.001). Thus, replacing HIV-1 infection status (Table 2) with CD4+ T cell count as a key non-genetic factor did not alter the independent associations of three HLA-DRB1 variants with Ab responses.
Analyses confined to 229 Hispanic subjects
In further analyses confined to 229 Hispanics (Table 5), a reduced multivariable model continued to confirm the negative impact of HIV-1 infection on Ab responses (P < 0.0001). DRB1*0804 (n = 11) and DRB1*15 (n = 47) were the only HLA class II variants independently associated with Ab responses (adjusted P = 0.045 and 0.019, respectively). Analyses of other alleles (including DRB1*03 and DRB1*0701) and their haplotypes were inconclusive because of reduced statistical power.
Secondary analyses of SNPs
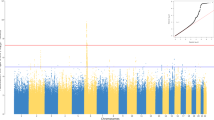
Secondary analyses tested the hypothesis that non-coding SNPs in and around HLA class II genes might also be contributory, because of their potential influence on gene expression and/or mRNA splicing, as implied by genome-wide association studies (Dalmasso et al. 2008; Fellay et al. 2007; Hafler et al. 2007; Kuniholm 2007; Limou et al. 2009). A panel of 88 SNPs within three blocks of human MHC was successfully genotyped in 280 patients with at least 2 μg of high-quality DNA at the completion of HLA genotyping. One SNP (rs2523685) mapped to MHC class I region had a nominal P value at 0.021 in test of SNP genotypes among the three Ab response groups (Fig. 1). This weak association was driven by the heterozygous genotype (rs2523685CG), which was present in 31 (37.8%) out of 82 HR, 70 (49.3%) out of 142 MR, and 33 (58.9%) out of 56 NR patients. Homozygosity for the minor allele (genotype GG) had no trend (24.4% HR, 14.1% MR, and 21.4% NR). The putative association of rs2523685 genotypes with differential Ab responses could be dismissed for three reasons. First, the rs2523685 SNP is mapped to the 5′ flanking region (distant from the promoter sequences) of HCP5 (Table S2), without any predicted or known functional relevance for either allele. Second, likelihood of chance association was high—about two random associations were expected from the tests of 88 SNPs when P value was accepted at 0.021. Alternatively, 1.3 random associations were expected from the tests of 62 MHC class I SNPs alone. Third, the SNP is remote from the HLA class II genes that produced the strongest signals in all analyses.
Distribution of nominal P values in the analyses of 88 single nucleotide polymorphisms (SNPs) within three regions of the human major histocompatibility complex (MHC) (Table S2). MHC class I SNPs (n = 62) are mapped to HLA class I and neighboring genes, including HCP5, HLA-C, and ZNRD1 previously associated with HIV-1-related outcomes (see text). MHC class III (central MHC) SNPs (n = 14) are mapped to TNF, LTA, and LTB, which encode TNF-α, lymphotoxic alpha, and lymphotoxic beta, respectively. MHC class II SNPs (n = 12) are in non-coding sequences around HLA-DRB1, -DQA1, and -DQB1. The nominal P values are based on Chi-square tests of non-responders (serum antibody concentrations <10 mIU/mL at week 28), responders (10–1,000 mIU/mL), and high responders (>1,000 mIU/mL). All SNPs are sorted by their relative order (from telomere to centromere) instead of actual distance on the short arm of chromosome 6. For clarity, three spaces are introduced between MHC blocks. Only a single SNP (rs2523685) in the 5′ flanking region of HCP5 has trend for association (P = 0.021)
Discussion
Our analyses of three HLA class II genes in 335 youth confirmed the negative impact of DRB1*03 and DRB1*0701 on peak Ab responses to HBV vaccination, as reported in prior studies of infant, adolescent, and adult populations (Belloni et al. 1993; Caillat-Zucman et al. 1998; Craven et al. 1986; del Canho et al. 1993; Hatae et al. 1992; Hohler et al. 1998, 2002b; Kramer et al. 1988; Martinetti et al. 1995; McDermott et al. 1997; Peces et al. 1997; Qian et al. 2002; Wang et al. 2004; Watanabe et al. 1988; Weissman et al. 1988). Although our study relied heavily on Hispanics and HIV+ youth, the main findings most closely resembled those reported earlier for adults of European ancestry (Hohler et al. 1998). Studies on patients with natural HBV infection lend further support for these immunogenetic findings, as HLA haplotypes having either DRB1*03 or DRB1*07 were enriched in patients who were unable to clear HBV after infection (Thio et al. 2003). In addition, as both DRB1*03 and DRB1*0701 are in strong LD with DQB1*02 (DQ2 by serology), their relationships to Ab responses were partially reflected by DQB1*02 (Belloni et al. 1993; Hohler et al. 1998; Martinetti et al. 1995, 2000; McDermott et al. 1997; Peces et al. 1997; Stachowski et al. 1995).
In population-based analyses of diverse HLA alleles, statistical power (sample size and allele frequency), false discoveries, and population heterogeneity are some of the major concerns. The HLA-DRB1 variants negatively associated with Ab responses were relatively common in our study population, but statistical power alone could not account for these key findings because statistical power actually favored the analyses of HLA-DQA1 and -DQB1 variants, which are much less diverse than HLA-DRB1 alleles. False discovery was also unlikely because various studies have led to similar conclusions. Indeed, even the minor finding on DRB1*15 (DR2 by serology), which appeared to be a favorable factor in HIV+ youth (Table 4), was also consistent with evidence from studies of infants and adults (Caillat-Zucman et al. 1993; Hohler et al. 1998). Population heterogeneity, on the other hand, could obscure causal relationships when analyses can only survey tagging variants instead of the functional units. However, heterogeneity in ethnic backgrounds could not compromise our major findings at least for three reasons. First, individual HLA alleles encode structurally and functionally distinct protein products that facilitate immune surveillance regardless of racial or ethnic background (Jones et al. 2006). Second, parameter estimates for DRB1*03 and DRB1*0701 were mostly insensitive to statistical adjustments for non-genetic factors, including race/ethnicity (Table 2). Third, our analyses dismissed the potential confounding by other genetic variants, especially those (e.g., DQB1*0201 and DQB1*0202) that are known to be in strong LD with DRB1*03 and DRB1*0701 (Table 2).
HLA class II alleles have been associated with differential Ab responses to other vaccines, including measles–mumps–rubella (MMR) vaccine (Ovsyannikova et al. 2004, 2006). In US schoolchildren (mostly European Americans) who received MMR vaccination, DRB1*08 was considered unfavorable—being negatively associated with desirable immune response (Ovsyannikova et al. 2004), which contrasted with its positive association with Ab response to HBV vaccination here. Another study reported a disadvantage for DRB1*0801 in primary biliary cirrhosis patients of European descent (Donaldson et al. 2006). While predominance of Hispanics and HIV+ subjects in our study population might be one of the reasons for seemingly inconsistent findings between studies, MMR vaccine is quite different from HBV vaccine in that it uses attenuated viruses instead of recombinant antigens. Moreover, DRB1*08 in our study population was represented by at least three common alleles (DRB1*0801, *0802, and *0804) and only DRB1*0804 had the clearest impact on Ab response to HBV vaccine (Table S2). In the absence of highly comparable data from other studies, the relevance of DRB1*08 to HBV vaccination deserves further evaluation in larger populations.
In contrast to HLA alleles with well-defined function, the MHC SNPs are mostly within non-coding sequences and are suitable for testing secondary hypotheses about relevance of gene expression and tagging of other HLA alleles (de Bakker et al. 2006; Listgarten et al. 2008; Miretti et al. 2005) not specifically captured in routine HLA genotyping. Our selection of SNPs in MHC class I region was dictated by recent studies, which have demonstrated the potential influence of MHC SNPs on HIV-1 pathogenesis and disease progression (Catano et al. 2008; Dalmasso et al. 2008; Fellay et al. 2007; Limou et al. 2009; Shrestha et al. 2009; van Manen et al. 2009). Although multiple SNPs can tag a few HLA class I alleles (e.g., B*5701) and serology groups (e.g., A10), these SNPs had little overlap with the panel of SNPs that can be used to reliably infer HLA class I alleles (Listgarten et al. 2008). The critical role of protective HLA class I alleles (e.g., B*5701 and B*5703) in immune control of HIV-1 infection (O’Brien and Nelson 2004; Tang and Kaslow 2003) was expected to indirectly benefit Ab response to HBV vaccination if patients are not receiving antiretroviral therapy. In analyses of MHC class I SNPs (Fig. 1), the absence of any apparent association was somewhat consistent with the earlier observation that HLA class I alleles and haplotypes were not associated with Ab responses in the REACH cohort (also consisted mostly of HIV-1-infected youth) (Wang et al. 2004). The new study population assembled here was not suitable for testing the indirect effects produced by HLA class I genotypes, because few HIV+ youth (less than 10%) had severe immunodeficiency at the time of vaccination. The weak signal produced by a single, non-coding SNP within the HCP5 gene is probably worth pursuing in larger, homogeneous cohorts, as analyses of SNPs without well-established functional attributes typically require stratification by race.
In summary, both primary and secondary analyses of HLA and related genotypes have reiterated multifactorial influences on Ab responses to HBV vaccination in youth. Recognition of immunodeficiency (the loss of CD4+ T cells) and several HLA-DRB1 alleles as independent predictors of Ab response highlights the importance of T-helper cell-dependent pathways and HLA class II antigen presentation to effective vaccination using recombinant antigens. This work has also paved the way for further high-throughput SNP typing that may reveal additional factors, including regulatory (non-coding) sequence variations commonly found in immune response genes beyond the HLA region, as well as coding sequence variations that are also common in genes encoding killer immunoglobulin receptors (Bashirova et al. 2006; Parham 2005). Systematic evaluation of non-HLA genes can be informative, as studies of European twins have already indicated that almost 59% of heritability in Ab responses to HBsAg can be attributed to genetic variations beyond the HLA-DRB1 locus (Hohler et al. 2002a).
References
Agrawal S, Khan F, Bharadwaj U (2007) Human genetic variation studies and HLA class II loci. Int J Immunogenet 34:247–252
Bashirova AA, Martin MP, McVicar DW, Carrington M (2006) The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet 7:277–300
Belloni C, Orsolini P, Martinetti M, Chirico G, Cerbo RM, Comolli G, Maccarini U, Barlassina C, Togni C, Polatti F et al (1993) Control of hepatitis B: evaluation of two different vaccinal schedules in newborns from HBsAg negative mothers. New Microbiol 16:237–244
Bergstrom TF, Erlandsson R, Engkvist H, Josefsson A, Erlich HA, Gyllensten U (1999) Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol Rev 167:351–365
Caillat-Zucman S, Gimenez JJ, Albouze G, Lebkiri B, Naret C, Jungers P, Bach JF (1993) HLA genetic heterogeneity of hepatitis B vaccine response in hemodialyzed patients. Kidney Int Suppl 41:S157–S160
Caillat-Zucman S, Gimenez JJ, Wambergue F, Albouze G, Lebkiri B, Naret C, Moynot A, Jungers P, Bach JF (1998) Distinct HLA class II alleles determine antibody response to vaccination with hepatitis B surface antigen. Kidney Int 53:1626–1630
Catano G, Kulkarni H, He W, Marconi VC, Agan BK, Landrum M, Anderson S, Delmar J, Telles V, Song L, Castiblanco J, Clark RA, Dolan MJ, Ahuja SK (2008) HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLoS ONE 3:e3636
Collier AC, Corey L, Murphy VL, Handsfield HH (1988) Antibody to human immunodeficiency virus (HIV) and suboptimal response to hepatitis B vaccination. Ann Intern Med 109:101–105
Cooper CL, Angel JB, Seguin I, Davis HL, Cameron DW (2008) CPG 7909 adjuvant plus hepatitis B virus vaccination in HIV-infected adults achieves long-term seroprotection for up to 5 years. Clin Infect Dis 46:1310–1314
Cornejo-Juarez P, Volkow-Fernandez P, Escobedo-Lopez K, Vilar-Compte D, Ruiz-Palacios G, Soto-Ramirez LE (2006) Randomized controlled trial of Hepatitis B virus vaccine in HIV-1-infected patients comparing two different doses. AIDS Res Ther 3:9
Craven DE, Awdeh ZL, Kunches LM, Yunis EJ, Dienstag JL, Werner BG, Polk BF, Syndman DR, Platt R, Crumpacker CS et al (1986) Nonresponsiveness to hepatitis B vaccine in health care workers. Results of revaccination and genetic typings. Ann Intern Med 105:356–360
Cruciani M, Mengoli C, Serpelloni G, Lanza A, Gomma M, Nardi S, Rimondo C, Bricolo F, Consolaro S, Trevisan M, Bosco O (2009) Serologic response to hepatitis B vaccine with high dose and increasing number of injections in HIV infected adult patients. Vaccine 27:17–22
Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV (2004) Global epidemiology of hepatitis B virus. J Clin Gastroenterol 38:S158–S168
Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, Chaix ML, Lambotte O, Avettand-Fenoel V, Le Clerc S, de Senneville LD, Deveau C, Boufassa F, Debre P, Delfraissy JF, Broet P, Theodorou I (2008) Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS ONE 3:e3907
de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD (2006) A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38:1166–1172
de Vries-Sluijs TE, Hansen BE, van Doornum GJ, Springeling T, Evertsz NM, de Man RA, van der Ende ME (2008) A prospective open study of the efficacy of high-dose recombinant hepatitis B rechallenge vaccination in HIV-infected patients. J Infect Dis 197:292–294
del Canho R, de Vries RR, Schalm SW (1993) Human leucocyte antigens (HLA) in neonates with an inadequate response to hepatitis B vaccination. Vaccine 11:983
Dienstag JL (2008) Hepatitis B virus infection. N Engl J Med 359:1486–1500
Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, Jones DE, James OF, Bassendine MF (2006) HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology 44:667–674
Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, Kaslow RA (2006) Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun 7:450–467
Fattovich G, Bortolotti F, Donato F (2008) Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol 48:335–352
Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB (2007) A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947
Fonseca MO, Pang LW, de Paula Cavalheiro N, Barone AA, Heloisa Lopes M (2005) Randomized trial of recombinant hepatitis B vaccine in HIV-infected adult patients comparing a standard dose to a double dose. Vaccine 23:2902–2908
Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL (2007) Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 357:851–862
Hatae K, Kimura A, Okubo R, Watanabe H, Erlich HA, Ueda K, Nishimura Y, Sasazuki T (1992) Genetic control of nonresponsiveness to hepatitis B virus vaccine by an extended HLA haplotype. Eur J Immunol 22:1899–1905
Hohjoh H, Ohashi J, Takasu M, Nishioka T, Ishida T, Tokunaga K (2003) Recent divergence of the HLA-DRB1*04 allelic lineage from the DRB1*0701 lineage after the separation of the human and chimpanzee species. Immunogenetics 54:856–861
Hohler T, Meyer CU, Notghi A, Stradmann-Bellinghausen B, Schneider PM, Starke R, Zepp F, Sanger R, Clemens R, Meyer zum Buschenfelde KH, Rittner C (1998) The influence of major histocompatibility complex class II genes and T-cell Vβ repertoire on response to immunization with HBsAg. Hum Immunol 59:212–218
Hohler T, Reuss E, Evers N, Dietrich E, Rittner C, Freitag CM, Vollmar J, Schneider PM, Fimmers R (2002a) Differential genetic determination of immune responsiveness to hepatitis B surface antigen and to hepatitis A virus: a vaccination study in twins. Lancet 360:991–995
Hohler T, Stradmann-Bellinghausen B, Starke R, Sanger R, Victor A, Rittner C, Schneider PM (2002b) C4A deficiency and nonresponse to hepatitis B vaccination. J Hepatol 37:387–392
Jones EY, Fugger L, Strominger JL, Siebold C (2006) MHC class II proteins and disease: a structural perspective. Nat Rev Immunol 6:271–282
Kramer A, Herth D, von Keyserlingk HJ, Ludwig WD, Hampl H, Sommer D, Hahn EG, Riecken EO (1988) Non-responsiveness to hepatitis-B vaccination: revaccination and immunogenetic typing. Klin Wochenschr 66:670–674
Kuniholm MH (2007) Replicating genome-wide association studies. Science 318:390–391, author reply 390–391
Lai CL, Yuen MF (2008) Chronic hepatitis B—new goals, new treatment. N Engl J Med 359:2488–2491
Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, Labib T, Taing L, Sladek R, Deveau C, Ratsimandresy R, Montes M, Spadoni JL, Lelievre JD, Levy Y, Therwath A, Schachter F, Matsuda F, Gut I, Froguel P, Delfraissy JF, Hercberg S, Zagury JF (2009) Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J Infect Dis 199:419–426
Listgarten J, Brumme Z, Kadie C, Xiaojiang G, Walker B, Carrington M, Goulder P, Heckerman D (2008) Statistical resolution of ambiguous HLA typing data. PLoS Comput Biol 4:e1000016
Martinetti M, Cuccia M, Daielli C, Ambroselli F, Gatti C, Pizzochero C, Belloni C, Orsolini P, Salvaneschi L (1995) Anti-HBV neonatal immunization with recombinant vaccine. Part II. Molecular basis of the impaired alloreactivity. Vaccine 13:555–560
Martinetti M, De Silvestri A, Belloni C, Pasi A, Tinelli C, Pistorio A, Salvaneschi L, Rondini G, Avanzini MA, Cuccia M (2000) Humoral response to recombinant hepatitis B virus vaccine at birth: role of HLA and beyond. Clin Immunol 97:234–240
McDermott AB, Zuckerman JN, Sabin CA, Marsh SG, Madrigal JA (1997) Contribution of human leukocyte antigens to the antibody response to hepatitis B vaccination. Tissue Antigens 50:8–14
McGovern BH (2007) The epidemiology, natural history and prevention of hepatitis B: implications of HIV coinfection. Antivir Ther 12(Suppl 3):H3–H13
Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, Morrison J, Whittaker P, Lander ES, Cardon LR, Bentley DR, Rioux JD, Beck S, Deloukas P (2005) A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 76:634–646
O’Brien SJ, Nelson GW (2004) Human genes that limit AIDS. Nat Genet 36:565–574
Ovsyannikova IG, Jacobson RM, Vierkant RA, Shane Pankratz V, Jacobsen SJ, Poland GA (2004) Associations between human leukocyte antigen (HLA) alleles and very high levels of measles antibody following vaccination. Vaccine 22:1914–1920
Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA (2006) Human leukocyte antigen haplotypes in the genetic control of immune response to measles–mumps–rubella vaccine. J Infect Dis 193:655–663
Parham P (2005) Influence of KIR diversity on human immunity. Adv Exp Med Biol 560:47–50
Pasricha N, Datta U, Chawla Y, Singh S, Arora SK, Sud A, Minz RW, Saikia B, Singh H, James I, Sehgal S (2006) Immune responses in patients with HIV infection after vaccination with recombinant Hepatitis B virus vaccine. BMC Infect Dis 6:65
Peces R, de la Torre M, Alcazar R, Urra JM (1997) Prospective analysis of the factors influencing the antibody response to hepatitis B vaccine in hemodialysis patients. Am J Kidney Dis 29:239–245
Qian Y, Zhang L, Liang XM, Hou JL, Luo KX (2002) Association of immune response to hepatitis B vaccine with HLA-DRB1*02, 07, 09 genes in the population of Han nationality in Guangdong Province. Di Yi Jun Yi Da Xue Xue Bao 22:67–69
Rey D, Krantz V, Partisani M, Schmitt MP, Meyer P, Libbrecht E, Wendling MJ, Vetter D, Nicolle M, Kempf-Durepaire G, Lang JM (2000) Increasing the number of hepatitis B vaccine injections augments anti-HBs response rate in HIV-infected patients. Effects on HIV-1 viral load. Vaccine 18:1161–1165
Robinson J, Waller MJ, Fail SC, Marsh SG (2006) The IMGT/HLA and IPD databases. Hum Mutat 27:1192–1199
Sasaki MG, Foccacia R, de Messias-Reason IJ (2003) Efficacy of granulocyte-macrophage colony-stimulating factor (GM-CSF) as a vaccine adjuvant for hepatitis B virus in patients with HIV infection. Vaccine 21:4545–4549
Schreuder GM, Hurley CK, Marsh SG, Lau M, Fernandez-Vina MA, Noreen HJ, Setterholm M, Maiers M (2005) HLA dictionary 2004: summary of HLA-A, -B, -C, -DRB1/3/4/5, -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Hum Immunol 66:170–210
Shao W, Tang J, Dorak MT, Song W, Lobashevsky E, Cobbs CS, Wrensch MR, Kaslow RA (2004) Molecular typing of human leukocyte antigen and related polymorphisms following whole genome amplification. Tissue Antigens 64:286–292
Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, Tang J (2009) Host genetics and HIV-1 viral set-point in African Americans. AIDS 23:673–677
Stachowski J, Kramer J, Fust G, Maciejewski J, Baldamus CA, Petranyi GG (1995) Relationship between the reactivity to hepatitis B virus vaccination and the frequency of MHC class I, II and III alleles in haemodialysis patients. Scand J Immunol 42:60–65
Tang J, Kaslow RA (2003) The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS 17:S51–S60
Tang J, Penman-Aguilar A, Lobashevsky E, Allen S, Kaslow R, Zambia-UAB HIV Research Project (2004) HLA-DRB1 and -DQB1 alleles and haplotypes in Zambian couples and their associations with heterosexual transmission of human immunodeficiency virus type 1. J Infect Dis 189:1696–1704
Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, Hunter E, Kaslow RA (2008) Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol 181:2626–2635
Thio CL, Thomas DL, Karacki P, Gao X, Marti D, Kaslow RA, Goedert JJ, Hilgartner M, Strathdee SA, Duggal P, O’Brien SJ, Astemborski J, Carrington M (2003) Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol 77:12083–12087
van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MAM, van’t Wout AB, Schuitemaker H (2009) Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS 23:19–28
Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA (2004) Human leukocyte antigen and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology 39:978–988
Watanabe H, Matsushita S, Kamikawaji N, Hirayama K, Okumura M, Sasazuki T (1988) Immune suppression gene on HLA-Bw54-DR4-DRw53 haplotype controls nonresponsiveness in humans to hepatitis B surface antigen via CD8+ suppressor T cells. Hum Immunol 22:9–17
Weissman JY, Tsuchiyose MM, Tong MJ, Co R, Chin K, Ettenger RB (1988) Lack of response to recombinant hepatitis B vaccine in nonresponders to the plasma vaccine. JAMA 260:1734–1738
Wilson CM, Ellenberg JH, Sawyer MK, Belzer M, Crowley-Nowick PA, Puga A, Futterman DC, Peralta L (2001) Serologic response to hepatitis B vaccine in HIV infected and high-risk HIV uninfected adolescents in the REACH cohort. Reaching for Excellence in Adolescent Care and Health. J Adolesc Health 29:123–129
Zuin G, Principi N, Tornaghi R, Paccagnini S, Re M, Massironi E, Ragni MC (1992) Impaired response to hepatitis B vaccine in HIV infected children. Vaccine 10:857–860
Acknowledgments
This work was supported by the Adolescent Trials Network for HIV/AIDS Interventions (ATN), with primary funding through two grants (U01 HD40474 and U01 HD40533) from National Institute of Child Health and Human Development (B. Kapogiannis, L. Serchuck) and supplemental funding from the National Institute of Allergy and Infectious Diseases (Patrick Jean-Philippe), National Institute on Drug Abuse (N. Borek), as well as National Institute of Mental Health (P. Brouwers, S. Allison). J.T. is the recipient of an independent scientist award (K02 AI076123) from National Institute of Allergy and Infectious Diseases. The entire project was scientifically reviewed by ATN’s Therapeutic Leadership Group. Network, scientific and logistical support was provided by the ATN Coordinating Center (C. Wilson, C. Partlow) at the University of Alabama at Birmingham. Network operations and analytic support came from the ATN Data and Operations Center at Westat Inc. (J. Korelitz, B. Driver). We sincerely thank members of the local youth Community Advisory Boards for their insight and counsel, the youth who participated in this study for their cooperation, Stephanie Crane (Adolescent Medicine Trials Unit) and Jenese Tucker (Westat) for valuable assistance with protocol implementation, Rick Mitchell (Westat) for data management, Richard A. Kaslow, Audrey Rogers, and Steven Spector for consultation and advice on protocol development, Karine Viaud (Illumina) for assistance with bioinformatics, Dale Isabelle and Mary Shirley for administrative assistance, Peter Havens, Sharon Nachman, Bret Rudy, Kathleen Squires, and John Sleasman for critical reading of an earlier version of this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
For the Adolescent Medicine Trials Network for HIV/AIDS Interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
This study (designated as ATN 052) also involved additional investigators and staff who enrolled patients at 22 clinical sites, including University of Maryland (Ligia Peralta, MD, Esther Collinetti, BA, Leonel Flores, MD, Reshma S. Gorle); Hospital dos Servidores do Estado, Rio de Janeiro (Esau C. Joao, MD, Leticia S. Cruz, MD, Eduarda Gusmao, MD, Angela B. N. Carvalho, MD); Universidade Federale de Minas Gerais, Minas Gerais (Jorge Pinto, MD, Flavia G. F. Ferreira, MD); St. Jude Research Hospital (Sarah Stender, MD, Kristen Branum, BS, Tina Culley, BS, RHIA, CCRP, Carla McKinley, RN, MSN, Thomas Wride, MS); Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto-Universidade de Sao Paulo, Sao Paulo (Geraldo Duarte, MD, Marisa M. M. Pinhata, MD, Carolina S. Vieira, MD); Instituto de Puericultura e Pediatria Martagao Geseteira, Rio de Janeiro (Ricardo H. S. Oliveira, MD, Maria C. C. Sapia, MD, Thalita F. Abreu, MD); Instituteo de Infectologia Emilio Ribas Sao Paulo (Marinella D. Negra, MD, Wladimir Queiroz, MD); Montefiore Medical Center (Donna Futterman, MD, Elizabeth Enriquez-Bruce, MD, Maria Campos, RN); Harriet Shezi Children’s Clinic, Johannesburg (Tammy Meyers, MD, Lee Kleynhans, MD, Megan Palmer, MD, Angela Oosthuizen, BS); John H. Stroger Jr. Hospital of Cook County and the CORE Center (Jaime Martinez, MD, Lisa Henry-Reid, Kelly Bojan, Rachel Jackson, MSN, APN, CFNP); University of Puerto Rico, Puerto Rico (Irma L. Febo, MD, Hazel T Ayala-Flores, Anne T. F. Gomez); Children’s Diagnostic and Treatment Center (Ana Puga, MD, Lisa Bridges, RN, Amy Inman, BS, Esmine Leonard, BSN); Children’s National Cancer Center (Lawrence J. D’Angelo, MD, Connie Trexler, RN, Rita Hagler, BA, Amy Klamberg, BA); Children’s Hospital of Los Angeles (Marvin Belzer, MD, Mirna Sweeney, RN, Diane Tucker, RN, MSN); Tulane University Health Sciences Center (Sue Ellen Abdalian, MD, Leslie Kozina, RN, CCRC, Alyne Baker, RN, MN, Trina Jeanjacques, BA); University of California at San Diego (Steven Spector, MD, Ronaldo M. Viani, MD, Jean M. Manning, BSN, Kim A. Norris, BSN, Lisa Stangl, PNP); University of South Florida (Patricia Emmanual, MD, Jorge Lujan-Zilbermann, MD, Diane M. Straub, MD, Silvia Callejas, BSN, ACRN, Priscilla C. Julian, RN, Amayvis Rebolledo, MAD); University of California at San Francisco (Babara Moscicki, MD, Coco Auerswald, MD, Lisa D. Irish, BSN, J. B. Molaghan, BA); Mount Sinai Medical Center (John B. Steever, MD, Angela Diaz, MD, Henry Sacks, MD, Mary S. Geiger, RN, MPH, Angela Lee, PA-C, MPH); Children’s Hospital of Boston (Cathryn Samples, MD, Brian Abascal, MFA, BA); Duke University (John Swetnam, MEd, Felicia Wiley, RN, Kareema Whitfield); Alabama Children’s Hospital (Robert Pass, MD, Heather Charlton, BSN, Beatty Newana, BA); San Juan City Hospital (Midnela Acevedo, MD, Milagros Gónzalez, MD, Lourdes Angeli, RN, MPH, Wanda Marrero, RN, Elvia Pérez, MEd, MA).
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Li, Y., Ni, R., Song, W. et al. Clear and independent associations of several HLA-DRB1 alleles with differential antibody responses to hepatitis B vaccination in youth. Hum Genet 126, 685–696 (2009). https://doi.org/10.1007/s00439-009-0720-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-009-0720-z